Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113599 |
| Name | oriT_S16BD03602|p105 |
| Organism | Salmonella enterica strain S16BD03602 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP099766 (14939..15024 [-], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | 61..68, 73..80 (TTGGTGGT..ACCACCAA) 27..34, 37..44 (GCAAAAAC..GTTTTTGC) 8..13, 21..26 (TGATTT..AAATCA) |
| Location of nic site | 53..54 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 86 nt
AATTACATGATTTAAAACGCAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file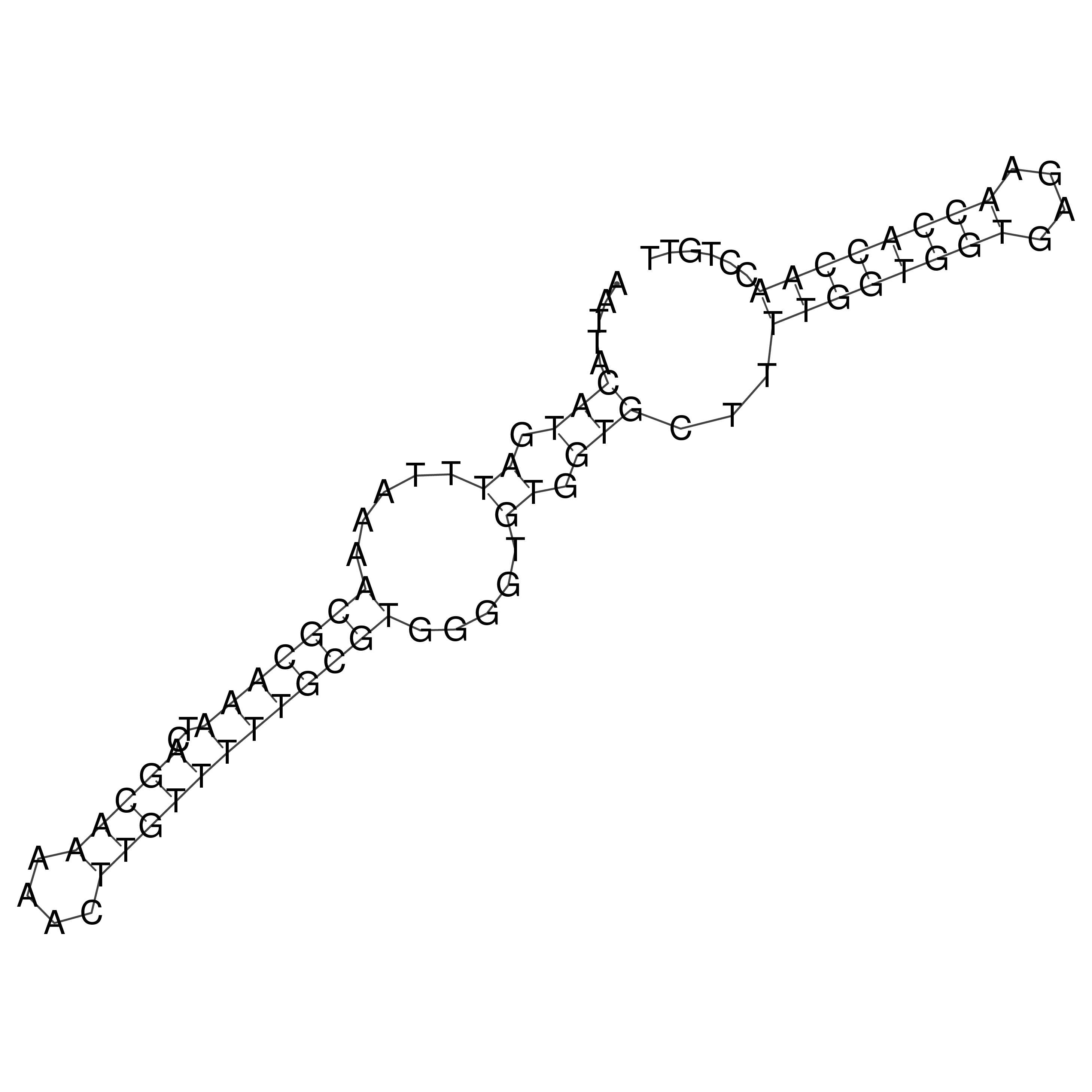
Relaxase
| ID | 8708 | GenBank | WP_253075075 |
| Name | traI_NHV93_RS00300_S16BD03602|p105 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191693.21 Da Isoelectric Point: 5.7446
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGKGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRTEIRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVVDSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPGSERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRLEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMQNGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWENNPDALALVDSVYHRIAGISKDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPRADREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVREIAGQERDRTATSEREAALPESVLRESQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 5091 | GenBank | WP_001342639 |
| Name | WP_001342639_S16BD03602|p105 |
UniProt ID | A0A836N7B7 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14508.47 Da Isoelectric Point: 4.7033
MARVILYISNDVYDKVNAIVEQRRQEGARDKDISLSGTASMLLELGLRVYEAQMERKESAFNQTEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A836N7B7 |
| ID | 5092 | GenBank | WP_000089263 |
| Name | WP_000089263_S16BD03602|p105 |
UniProt ID | A0A3U3XJL9 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 8541.74 Da Isoelectric Point: 8.6796
MSRNIIRPAPGNKVLLVLDDATNHKLLGARERSGRTKTNEVLVRLRDHLNRFPDFYNLDAIKEGAEETDS
IIKDL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3U3XJL9 |
T4CP
| ID | 10082 | GenBank | WP_000069752 |
| Name | traC_NHV93_RS00195_S16BD03602|p105 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99572.55 Da Isoelectric Point: 6.6265
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAA
ATQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASFHGAKITTQTVDAQAFIEIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPEMSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFKGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKKKQARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQY
TANGTYGRYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRTLKKLNVIDEGW
RLLDFKNRKVGEFIQKGYRTCRRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLFPDQFQPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRREG
MSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10083 | GenBank | WP_048961038 |
| Name | traD_NHV93_RS00295_S16BD03602|p105 |
UniProt ID | _ |
| Length | 726 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 726 a.a. Molecular weight: 82489.07 Da Isoelectric Point: 5.0504
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPDIQQ
QMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 14371..42330
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NHV93_RS00065 | 9755..10009 | + | 255 | WP_001230709 | hypothetical protein | - |
| NHV93_RS00070 | 10006..10248 | + | 243 | WP_000151456 | hypothetical protein | - |
| NHV93_RS00075 | 10430..10663 | + | 234 | WP_000553848 | hypothetical protein | - |
| NHV93_RS00080 | 10795..11088 | + | 294 | WP_112367531 | hypothetical protein | - |
| NHV93_RS00085 | 11396..11683 | + | 288 | WP_000606467 | hypothetical protein | - |
| NHV93_RS00090 | 12111..12344 | + | 234 | WP_000093745 | hypothetical protein | - |
| NHV93_RS00095 | 12420..12722 | + | 303 | WP_000379301 | hypothetical protein | - |
| NHV93_RS00100 | 12994..13143 | + | 150 | WP_000903193 | hypothetical protein | - |
| NHV93_RS00105 | 13254..14075 | + | 822 | WP_073972859 | DUF932 domain-containing protein | - |
| NHV93_RS00110 | 14371..14973 | - | 603 | WP_072205450 | transglycosylase SLT domain-containing protein | virB1 |
| NHV93_RS00115 | 15296..15679 | + | 384 | WP_001342639 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| NHV93_RS00120 | 15870..16523 | + | 654 | WP_001342640 | conjugal transfer protein TraJ | - |
| NHV93_RS00125 | 16648..16875 | + | 228 | WP_000089263 | conjugal transfer relaxosome protein TraY | - |
| NHV93_RS00130 | 16908..17270 | + | 363 | WP_023910486 | type IV conjugative transfer system pilin TraA | - |
| NHV93_RS00135 | 17275..17586 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| NHV93_RS00140 | 17608..18174 | + | 567 | WP_000399805 | type IV conjugative transfer system protein TraE | traE |
| NHV93_RS00145 | 18161..18889 | + | 729 | WP_001230809 | type-F conjugative transfer system secretin TraK | traK |
| NHV93_RS00150 | 18889..20316 | + | 1428 | WP_032215882 | F-type conjugal transfer pilus assembly protein TraB | traB |
| NHV93_RS00155 | 20306..20890 | + | 585 | WP_000002795 | conjugal transfer pilus-stabilizing protein TraP | - |
| NHV93_RS00160 | 20877..21197 | + | 321 | WP_001057302 | conjugal transfer protein TrbD | virb4 |
| NHV93_RS00165 | 21190..21441 | + | 252 | WP_001038341 | conjugal transfer protein TrbG | - |
| NHV93_RS00170 | 21438..21953 | + | 516 | WP_000809891 | type IV conjugative transfer system lipoprotein TraV | traV |
| NHV93_RS00175 | 22088..22309 | + | 222 | WP_001278978 | conjugal transfer protein TraR | - |
| NHV93_RS00180 | 22302..22778 | + | 477 | WP_001744739 | hypothetical protein | - |
| NHV93_RS00185 | 22858..23076 | + | 219 | WP_072277258 | hypothetical protein | - |
| NHV93_RS00190 | 23104..23451 | + | 348 | WP_160540375 | hypothetical protein | - |
| NHV93_RS00195 | 23577..26207 | + | 2631 | WP_000069752 | type IV secretion system protein TraC | virb4 |
| NHV93_RS00200 | 26204..26590 | + | 387 | WP_000214082 | type-F conjugative transfer system protein TrbI | - |
| NHV93_RS00205 | 26587..27219 | + | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| NHV93_RS00210 | 27216..28208 | + | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| NHV93_RS00215 | 28238..28543 | + | 306 | WP_000224411 | hypothetical protein | - |
| NHV93_RS00220 | 28552..29190 | + | 639 | WP_032299189 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| NHV93_RS00225 | 29187..29558 | + | 372 | WP_000056340 | hypothetical protein | - |
| NHV93_RS00230 | 29583..30005 | + | 423 | WP_001229306 | HNH endonuclease | - |
| NHV93_RS00235 | 30002..31810 | + | 1809 | WP_237828321 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| NHV93_RS00240 | 31837..32094 | + | 258 | WP_000864319 | conjugal transfer protein TrbE | - |
| NHV93_RS00245 | 32087..32830 | + | 744 | WP_001030371 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| NHV93_RS00250 | 32846..32944 | + | 99 | Protein_49 | conjugal transfer protein TrbA | - |
| NHV93_RS00255 | 32998..33282 | + | 285 | WP_000623730 | type-F conjugative transfer system pilin chaperone TraQ | - |
| NHV93_RS00260 | 33269..33814 | + | 546 | WP_000059832 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| NHV93_RS00265 | 33744..34091 | + | 348 | WP_072130275 | P-type conjugative transfer protein TrbJ | - |
| NHV93_RS00270 | 34072..34464 | + | 393 | WP_001462107 | F-type conjugal transfer protein TrbF | - |
| NHV93_RS00275 | 34451..35824 | + | 1374 | WP_088130027 | conjugal transfer pilus assembly protein TraH | traH |
| NHV93_RS00280 | 35821..38646 | + | 2826 | WP_061089748 | conjugal transfer mating-pair stabilization protein TraG | traG |
| NHV93_RS00285 | 38643..39152 | + | 510 | WP_160540419 | conjugal transfer entry exclusion protein TraS | - |
| NHV93_RS00290 | 39166..39897 | + | 732 | WP_001734698 | conjugal transfer complement resistance protein TraT | - |
| NHV93_RS00295 | 40150..42330 | + | 2181 | WP_048961038 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 14034 | GenBank | NZ_CP099766 |
| Plasmid name | S16BD03602|p105 | Incompatibility group | IncFII |
| Plasmid size | 52174 bp | Coordinate of oriT [Strand] | 14939..15024 [-] |
| Host baterium | Salmonella enterica strain S16BD03602 |
Cargo genes
| Drug resistance gene | blaCTX-M-15 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |