Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113595 |
| Name | oriT_pT173a |
| Organism | Ensifer canadensis strain T173 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP083375 (88260..88297 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pT173a
GGATCCAAGGGCGCAATTATACGTCGCTGATGCGACGC
GGATCCAAGGGCGCAATTATACGTCGCTGATGCGACGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file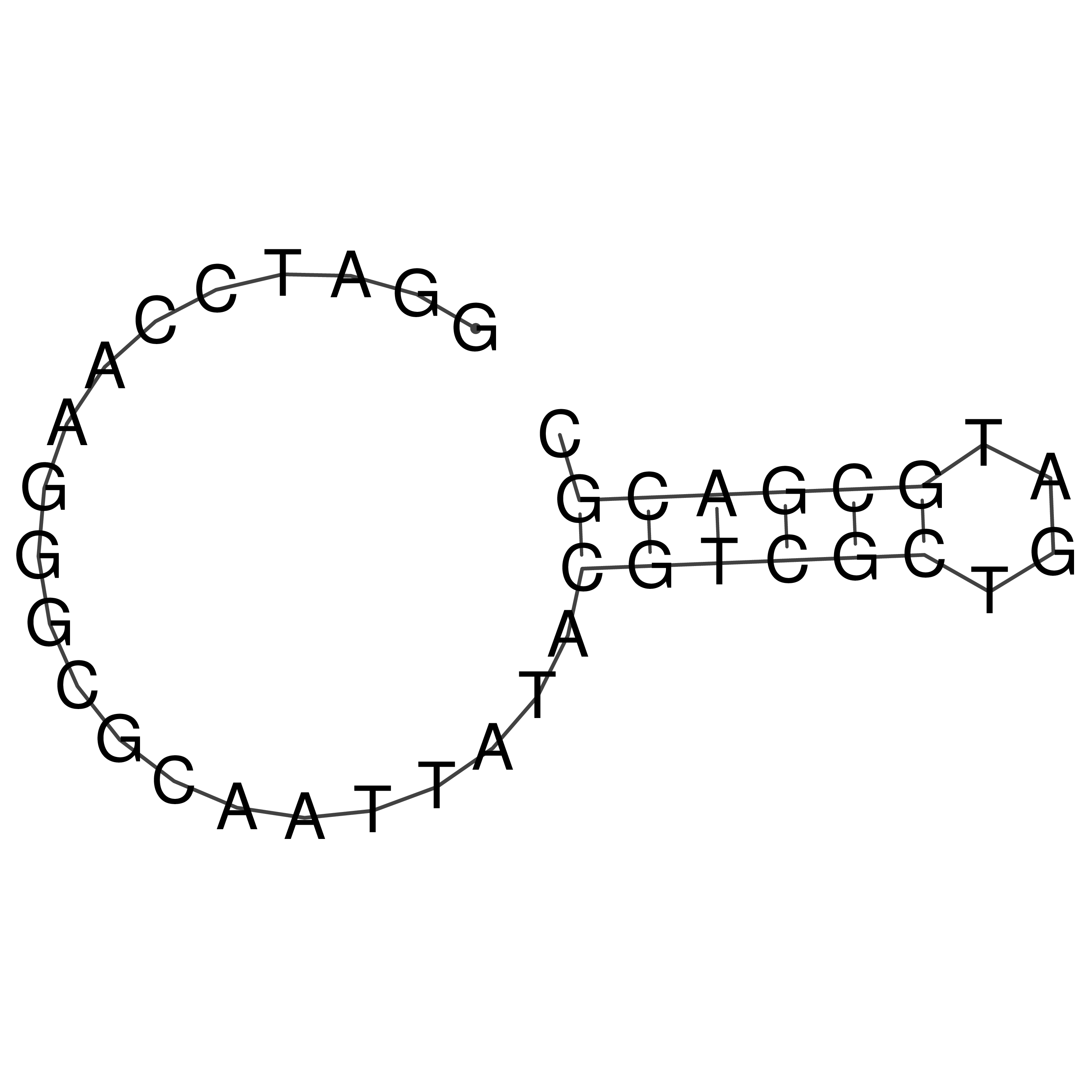
Relaxase
| ID | 8705 | GenBank | WP_203529550 |
| Name | traA_J3R84_RS38090_pT173a |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123110.58 Da Isoelectric Point: 10.0731
>WP_203529550.1 Ti-type conjugative transfer relaxase TraA [Ensifer canadensis]
MAIAHFSVNIVSRGDGRSAVLSAAYRHCAKMDYEREARTIDYSRKEGLLHEEFVLPADAPQWARKLIADS
SVSGASEAFWNKVEAFEKRIDAQLAKEVTIALPLELKPKQNVALVRDFVEKHILSQGMAADWVYHDNPGN
PHIHLMTTLRPLTEEGFGPKKVAVIGEDGQPLRNTAGKIVYELWAGGNKDFNAFRDAWFERQNHHLTLNG
VALQVDGRSYEKQGIELRPTIHLGVGAKAIERKAVVEGERPSLERLDLQEARKTENVRRIQARPEIVLDL
VSREKSVFDERDVAKVLHRYIDDQGIFQQLLARILQSPEILRLQRDTIEFATGQRLPARYTTRELIRLEA
EMARRSLWLSERKTHGVSKTVLEATFARHARLSAEQRVAIEHVAGSARVAAVVGRAGAGKTSMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWKKDRDTLDANTVFVMDEAGMVSSRQMAGFVE
AAVKSGAKLVLVGDPEQLQPIEAGAAFRAIADRVGYAELETIYRQREDWMRKASLDLARGRVGEALIAYR
SEGKVLGAELKAQAVEHLVADWNRDYDPSKSMLMLAHLRRDVRMLNVMAREKLVERGILGEGHSFRTADG
IRHFDAGDQIVFLKNEGSLGVKNGMIGRVVEAAPNRISVVIGEGDQRRQVSVEQRFYNNLDHGYATTVHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLALYYGRRSFSKAGGLTQILSRRNAKETTLDYERGNLY
RPALAFAENRGLHIVQVARTLLRDRLDWTLRQSSKLADLAVRLRAAGERLGLLQTLKTQTTKETRPMVSG
VKLFPVLLSDVVGRKVADDPAVKKQWDEVSTRFRYVFADPETAFRTMNFDAVLADSQVASQTLNKLAADP
SSIGPLKGKTGVLASKSDREARRVAEVNVPALRRDIETYLRIREVAVQRLESEEKTQRLRVSIDIPALSP
AAQSVLERVRDAIDRSDLPTAMAYALSNRETKAEIDGFGKAVTERFGERTLLSNAARDPQGQLFDKLSEG
LAPQEKERLKEAWPVMRTAQQLSAHERTVVTLKQVEDLKLAQRQTPMIKQ
MAIAHFSVNIVSRGDGRSAVLSAAYRHCAKMDYEREARTIDYSRKEGLLHEEFVLPADAPQWARKLIADS
SVSGASEAFWNKVEAFEKRIDAQLAKEVTIALPLELKPKQNVALVRDFVEKHILSQGMAADWVYHDNPGN
PHIHLMTTLRPLTEEGFGPKKVAVIGEDGQPLRNTAGKIVYELWAGGNKDFNAFRDAWFERQNHHLTLNG
VALQVDGRSYEKQGIELRPTIHLGVGAKAIERKAVVEGERPSLERLDLQEARKTENVRRIQARPEIVLDL
VSREKSVFDERDVAKVLHRYIDDQGIFQQLLARILQSPEILRLQRDTIEFATGQRLPARYTTRELIRLEA
EMARRSLWLSERKTHGVSKTVLEATFARHARLSAEQRVAIEHVAGSARVAAVVGRAGAGKTSMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWKKDRDTLDANTVFVMDEAGMVSSRQMAGFVE
AAVKSGAKLVLVGDPEQLQPIEAGAAFRAIADRVGYAELETIYRQREDWMRKASLDLARGRVGEALIAYR
SEGKVLGAELKAQAVEHLVADWNRDYDPSKSMLMLAHLRRDVRMLNVMAREKLVERGILGEGHSFRTADG
IRHFDAGDQIVFLKNEGSLGVKNGMIGRVVEAAPNRISVVIGEGDQRRQVSVEQRFYNNLDHGYATTVHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLALYYGRRSFSKAGGLTQILSRRNAKETTLDYERGNLY
RPALAFAENRGLHIVQVARTLLRDRLDWTLRQSSKLADLAVRLRAAGERLGLLQTLKTQTTKETRPMVSG
VKLFPVLLSDVVGRKVADDPAVKKQWDEVSTRFRYVFADPETAFRTMNFDAVLADSQVASQTLNKLAADP
SSIGPLKGKTGVLASKSDREARRVAEVNVPALRRDIETYLRIREVAVQRLESEEKTQRLRVSIDIPALSP
AAQSVLERVRDAIDRSDLPTAMAYALSNRETKAEIDGFGKAVTERFGERTLLSNAARDPQGQLFDKLSEG
LAPQEKERLKEAWPVMRTAQQLSAHERTVVTLKQVEDLKLAQRQTPMIKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10078 | GenBank | WP_203529556 |
| Name | traG_J3R84_RS38075_pT173a |
UniProt ID | _ |
| Length | 656 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 656 a.a. Molecular weight: 71263.93 Da Isoelectric Point: 9.3488
>WP_203529556.1 Ti-type conjugative transfer system protein TraG [Ensifer canadensis]
MTVNRLLLTLLPVTIMVAVIVLMSGVELWLAAFGKTAQAKLMLGRIGLALPYVTAAAIGVIFLFAANGAE
NIKAAGWGVVIGCVATILIAVLHEATRLANIADIVPSDQSVLAYADPATMLGAATVFLAGVFAMRVVLRG
NAAFADAAPRRIRGKRAVHGEADWMSMVESEKLFPDAGGIVIGERYRVDRESVSRVAFRADRRETWGAGG
KTPLLCFDGSFGSSHGIVFAGSGGYKTTSVTIPTALKWGGGLVVLDPSSEVAPMVVDHRRRAGRKVIVLD
AADPTTGFNALDWIGRFGATREEDIVAVATWIMTDNARAASARDDFFRASAMQLLTALIADVCLSGHTDA
TDQTLRRVRANLSEPEPKLRERLTRIYEQSESTFVRENVAVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGDSFSTDELATGVTDIFIALDLKVLESHPGLARVVIGSLLNAIYNRNGEVAGRTLFLLDEVARLGY
LRILETARDAGRKYGISLALIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPETAEYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLTRRPLILPHEVMRMRGDEQIVFTAGNAPLRCGRAIWFRRADMISIVG
VNRFHRTAPEREAVSKAETPLTPLRK
MTVNRLLLTLLPVTIMVAVIVLMSGVELWLAAFGKTAQAKLMLGRIGLALPYVTAAAIGVIFLFAANGAE
NIKAAGWGVVIGCVATILIAVLHEATRLANIADIVPSDQSVLAYADPATMLGAATVFLAGVFAMRVVLRG
NAAFADAAPRRIRGKRAVHGEADWMSMVESEKLFPDAGGIVIGERYRVDRESVSRVAFRADRRETWGAGG
KTPLLCFDGSFGSSHGIVFAGSGGYKTTSVTIPTALKWGGGLVVLDPSSEVAPMVVDHRRRAGRKVIVLD
AADPTTGFNALDWIGRFGATREEDIVAVATWIMTDNARAASARDDFFRASAMQLLTALIADVCLSGHTDA
TDQTLRRVRANLSEPEPKLRERLTRIYEQSESTFVRENVAVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGDSFSTDELATGVTDIFIALDLKVLESHPGLARVVIGSLLNAIYNRNGEVAGRTLFLLDEVARLGY
LRILETARDAGRKYGISLALIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPETAEYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLTRRPLILPHEVMRMRGDEQIVFTAGNAPLRCGRAIWFRRADMISIVG
VNRFHRTAPEREAVSKAETPLTPLRK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10079 | GenBank | WP_203529997 |
| Name | t4cp2_J3R84_RS38300_pT173a |
UniProt ID | _ |
| Length | 820 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 820 a.a. Molecular weight: 91646.28 Da Isoelectric Point: 5.7567
>WP_203529997.1 conjugal transfer protein TrbE [Ensifer canadensis]
MVALKPFRHSGPSFADLVPYAGLVDNGVVLLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINTILSRLG
TGWMIQVEAVRVPTIDYPAEDVCHFPDPVTRAIDAERRSHFERESGHFESRHALVLTWRPPEPRRSGLAR
YVYADAGSRSATYAETALETFRTSIREVEQYLANVISIHRMVTHEAEDRGGFRIARYDELFQFVRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLAPRVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDEQEARAKLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIIL
FDEDSTRLQEKCEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYANIREPLINTRNLADLVPLNSVWS
GSPTAPCPFYPTGSPPLMQVASGSTPFRLNLHVGDVGHTLVFGPTGSGKSTLLALIAAQFRRYADGQIFA
FDKGRSMLPLTLAGGGDHYEIGGESDGKGRALAFCPLSDLSTDADRAWATEWIETLVALQGVAITPDHRN
AISRQIGLMAEARGRSLSDFVSGVQMREIKEALHHYTIDGPMGQLLDAEEDGLALGAFQTFELEELMNMG
ERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWIMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIVDVLKESCPTKICLPNGAARETGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDMTLGP
VALSFVGATGKDDLARVRALETQHVAEWPLHWLEQRGIAHAARYFPATKV
MVALKPFRHSGPSFADLVPYAGLVDNGVVLLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINTILSRLG
TGWMIQVEAVRVPTIDYPAEDVCHFPDPVTRAIDAERRSHFERESGHFESRHALVLTWRPPEPRRSGLAR
YVYADAGSRSATYAETALETFRTSIREVEQYLANVISIHRMVTHEAEDRGGFRIARYDELFQFVRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLAPRVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDEQEARAKLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIIL
FDEDSTRLQEKCEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYANIREPLINTRNLADLVPLNSVWS
GSPTAPCPFYPTGSPPLMQVASGSTPFRLNLHVGDVGHTLVFGPTGSGKSTLLALIAAQFRRYADGQIFA
FDKGRSMLPLTLAGGGDHYEIGGESDGKGRALAFCPLSDLSTDADRAWATEWIETLVALQGVAITPDHRN
AISRQIGLMAEARGRSLSDFVSGVQMREIKEALHHYTIDGPMGQLLDAEEDGLALGAFQTFELEELMNMG
ERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWIMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIVDVLKESCPTKICLPNGAARETGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDMTLGP
VALSFVGATGKDDLARVRALETQHVAEWPLHWLEQRGIAHAARYFPATKV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 123949..133590
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| J3R84_RS38235 (J3R84_37960) | 119208..119852 | + | 645 | WP_225968614 | lantibiotic dehydratase C-terminal domain-containing protein | - |
| J3R84_RS38240 | 120082..120363 | - | 282 | WP_225906554 | 3'-5' exonuclease | - |
| J3R84_RS38245 (J3R84_37970) | 120409..120783 | + | 375 | WP_203529980 | hypothetical protein | - |
| J3R84_RS38250 (J3R84_37975) | 120927..121025 | - | 99 | Protein_98 | AAA family ATPase | - |
| J3R84_RS38255 (J3R84_37980) | 121074..121785 | + | 712 | Protein_99 | IS6 family transposase | - |
| J3R84_RS38260 (J3R84_37985) | 121930..122898 | - | 969 | WP_203529981 | acyltransferase domain-containing protein | - |
| J3R84_RS38265 (J3R84_37990) | 122918..123913 | - | 996 | WP_203529982 | acyltransferase domain-containing protein | - |
| J3R84_RS38270 (J3R84_37995) | 123949..125250 | - | 1302 | WP_203529983 | IncP-type conjugal transfer protein TrbI | virB10 |
| J3R84_RS38275 (J3R84_38000) | 125262..125711 | - | 450 | WP_203529984 | conjugal transfer protein TrbH | - |
| J3R84_RS38280 (J3R84_38005) | 125711..126556 | - | 846 | WP_203529985 | P-type conjugative transfer protein TrbG | virB9 |
| J3R84_RS38285 (J3R84_38010) | 126574..127239 | - | 666 | WP_203529986 | conjugal transfer protein TrbF | virB8 |
| J3R84_RS38290 (J3R84_38015) | 127270..128466 | - | 1197 | WP_203529987 | P-type conjugative transfer protein TrbL | virB6 |
| J3R84_RS38985 (J3R84_38020) | 128460..128690 | - | 231 | WP_203529988 | entry exclusion protein TrbK | - |
| J3R84_RS38295 (J3R84_38025) | 128684..129460 | - | 777 | WP_203529989 | P-type conjugative transfer protein TrbJ | virB5 |
| J3R84_RS38300 (J3R84_38030) | 129465..131927 | - | 2463 | WP_203529997 | conjugal transfer protein TrbE | virb4 |
| J3R84_RS38305 (J3R84_38035) | 131930..132229 | - | 300 | WP_203529990 | conjugal transfer protein TrbD | virB3 |
| J3R84_RS38310 (J3R84_38040) | 132222..132629 | - | 408 | WP_203529991 | conjugal transfer pilin TrbC | - |
| J3R84_RS38315 (J3R84_38045) | 132619..133590 | - | 972 | WP_203529992 | P-type conjugative transfer ATPase TrbB | virB11 |
| J3R84_RS38320 (J3R84_38050) | 133587..134222 | - | 636 | WP_203529993 | acyl-homoserine-lactone synthase | - |
| J3R84_RS38325 (J3R84_38055) | 134583..135797 | + | 1215 | WP_203529994 | plasmid partitioning protein RepA | - |
| J3R84_RS38330 (J3R84_38060) | 135824..136834 | + | 1011 | WP_225906555 | plasmid partitioning protein RepB | - |
| J3R84_RS38335 (J3R84_38065) | 137076..138356 | + | 1281 | WP_207932912 | plasmid replication protein RepC | - |
Host bacterium
| ID | 14030 | GenBank | NZ_CP083375 |
| Plasmid name | pT173a | Incompatibility group | - |
| Plasmid size | 195834 bp | Coordinate of oriT [Strand] | 88260..88297 [+] |
| Host baterium | Ensifer canadensis strain T173 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | katB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |