Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113496 |
| Name | oriT_17YN198|p1 |
| Organism | Leclercia adecarboxylata strain 17YN198 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP106960 (48189..48272 [+], 84 nt) |
| oriT length | 84 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 84 nt
>oriT_17YN198|p1
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTAATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTTTT
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTAATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file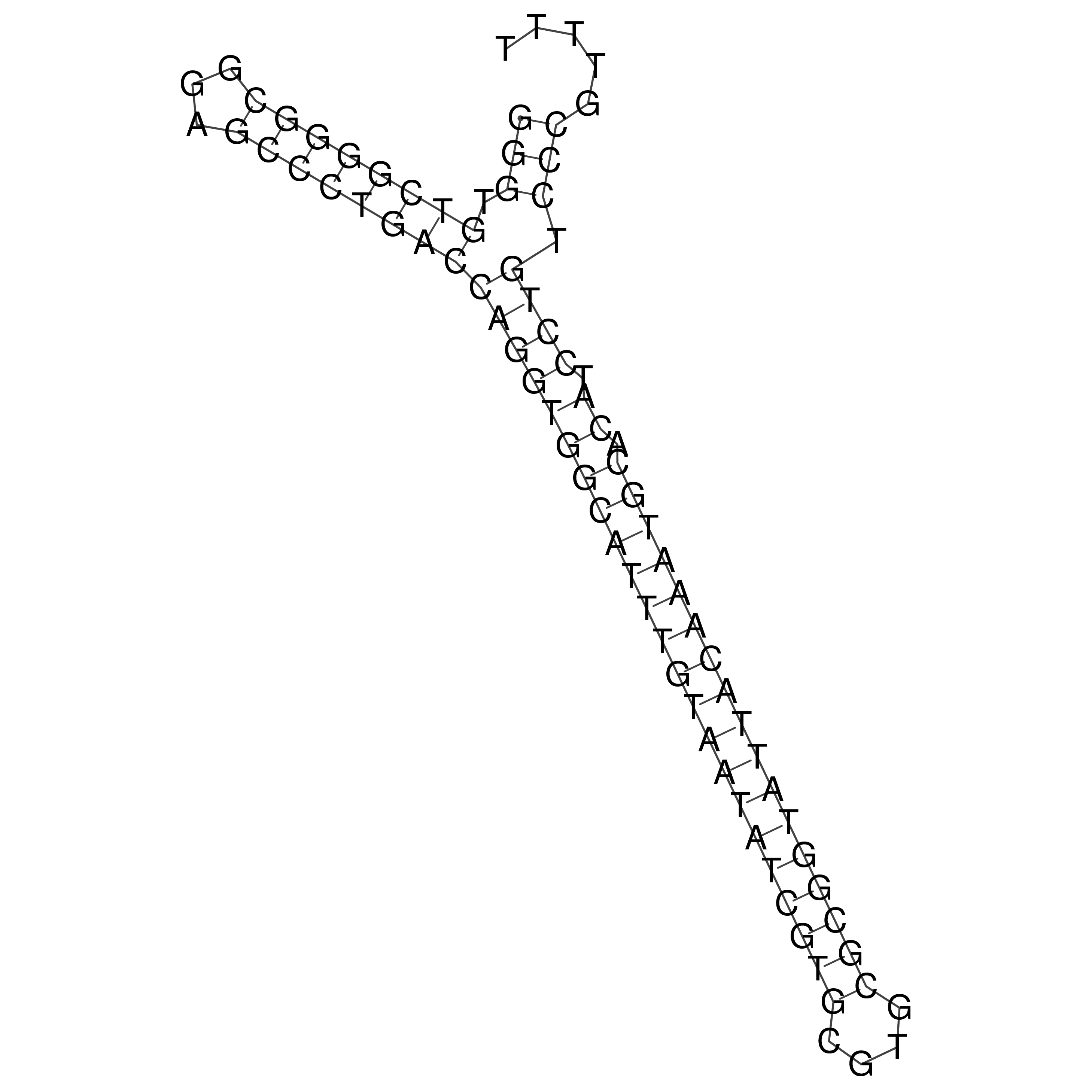
Relaxase
| ID | 8646 | GenBank | WP_276517572 |
| Name | Relaxase_OCT50_RS22355_17YN198|p1 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 102671.62 Da Isoelectric Point: 8.4473
>WP_276517572.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MIPVIPKKRKDGKSSFGDLVSYVSVRDEPQDDDLMEAVRASGTKADMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFAARKAKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVRHSLSRLGLSDHQFVSAVHTDTDNLHVHVAVNRVHPQTGYLNRLPYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSVERDRQSAWQHGRKPSLKEYIADTAVAGLREAPVTDWPSLHQRFAQAGLFIASD
NGELKVKDGWDRERPGVTLSAFGHSWNTEKLIRKLGEFTPPAQDIFTQVPDVGRYDPDTVTMPSRPERMS
EKETLNEYAVSRLRAPLIALDQNPAQRTMQAVHALLAHSGLYLKEQHDRLVICDGYDQTRTPVRAERVWP
ALTKAVMNSYEGGWQPVPHDIFRQVPPAERFAGGGLEARPVSDREWAKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDLEALISSGEFTWERGHELLARQGLLLMREQQGLMVMDAYNHDQTPVKASHVHPDLTLAR
AEPHAGAFVRVPSDIFERVAPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAARDDLKARYSAWRAN
WQRPDLRNRERWQAIHDECRLRKARIRAQQRDPLVRKLHYHIAELQRMQALIALKESVKAERLELIGQGK
WYPPSYRQWVEVEALKGDKAAISQLRGWDYRDRRSNAVKATTDNRCVVICEPGGTPVYSGVPGLRASLKK
NGRVQFRDPETGRHICTDYGDRVIFQNNGDYDVLKHDMVKVAPVLFSRSPTVAMEPEGNNDQFNAAFAKM
VAWHNVNHRDEGEYQISRPDVDQLRVESERRFDEMVNQGIHTLDNNVAYQNTWKPPSPR
MIPVIPKKRKDGKSSFGDLVSYVSVRDEPQDDDLMEAVRASGTKADMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFAARKAKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVRHSLSRLGLSDHQFVSAVHTDTDNLHVHVAVNRVHPQTGYLNRLPYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSVERDRQSAWQHGRKPSLKEYIADTAVAGLREAPVTDWPSLHQRFAQAGLFIASD
NGELKVKDGWDRERPGVTLSAFGHSWNTEKLIRKLGEFTPPAQDIFTQVPDVGRYDPDTVTMPSRPERMS
EKETLNEYAVSRLRAPLIALDQNPAQRTMQAVHALLAHSGLYLKEQHDRLVICDGYDQTRTPVRAERVWP
ALTKAVMNSYEGGWQPVPHDIFRQVPPAERFAGGGLEARPVSDREWAKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDLEALISSGEFTWERGHELLARQGLLLMREQQGLMVMDAYNHDQTPVKASHVHPDLTLAR
AEPHAGAFVRVPSDIFERVAPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAARDDLKARYSAWRAN
WQRPDLRNRERWQAIHDECRLRKARIRAQQRDPLVRKLHYHIAELQRMQALIALKESVKAERLELIGQGK
WYPPSYRQWVEVEALKGDKAAISQLRGWDYRDRRSNAVKATTDNRCVVICEPGGTPVYSGVPGLRASLKK
NGRVQFRDPETGRHICTDYGDRVIFQNNGDYDVLKHDMVKVAPVLFSRSPTVAMEPEGNNDQFNAAFAKM
VAWHNVNHRDEGEYQISRPDVDQLRVESERRFDEMVNQGIHTLDNNVAYQNTWKPPSPR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 5031 | GenBank | WP_231070074 |
| Name | WP_231070074_17YN198|p1 |
UniProt ID | _ |
| Length | 111 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 111 a.a. Molecular weight: 12726.54 Da Isoelectric Point: 10.3917
>WP_231070074.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSDRASRRGSEKRQKTVIKTLRFTPEEIEQIEREAEQAGQSVSAFIRNAAMNRKVIPAIDDSFLTELLRL
GRLQKHHFVEGKRVGDKEYSTVLVAITELANTLRKQLMNND
MSDRASRRGSEKRQKTVIKTLRFTPEEIEQIEREAEQAGQSVSAFIRNAAMNRKVIPAIDDSFLTELLRL
GRLQKHHFVEGKRVGDKEYSTVLVAITELANTLRKQLMNND
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 9993 | GenBank | WP_241281505 |
| Name | trbC_OCT50_RS22345_17YN198|p1 |
UniProt ID | _ |
| Length | 765 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 765 a.a. Molecular weight: 87123.29 Da Isoelectric Point: 6.3935
>WP_241281505.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MNNTPVNQAMMKRSAWQSPLLEMLRDHLLYAGIMAGSLVAGFIWPLAIPLLLFLAIIASVSFSTHRWRMP
MRMPVHLDRVDPSEDRKTRRSLFSAFPTLVQYEAIQERKGRGIFYLGYQRMRDTGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IQNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKNMSL
QLLREFMPLEKLAGLYCRAVDDQWPEEAVAPLYNYLVDVPGFDMSMVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHKLE
GLDSEALEITKYKGAFPYMNYLDEVGAYYTERIAVQATQVRSLDFSIIMMSQDQERIESQTSATNVATLM
QNAGTKVAGKIVSDDKTAKTISNAAGQEARANMVSLQRQDGLIGTSWIDGNQINIQMENKINVQDLITLQ
PGENYTVFQGDPVPGASFYIEPEDKTCGEPLIINRYITINPPGLKQLRKLVPRTAQRRLPTPENVSKIIG
VLTAKPSRRKKKSGGEPWKVIDTFQQRLANRQEAHNLMTEYDMDHSGREKSLWEEALEVIRSTTMAERTI
RYITLNRPEPEPSEPGDEENSQPQEVILKNLTLPAPVIRPARTGNEPPQQISPHIPLRPEMEWPE
MNNTPVNQAMMKRSAWQSPLLEMLRDHLLYAGIMAGSLVAGFIWPLAIPLLLFLAIIASVSFSTHRWRMP
MRMPVHLDRVDPSEDRKTRRSLFSAFPTLVQYEAIQERKGRGIFYLGYQRMRDTGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IQNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKNMSL
QLLREFMPLEKLAGLYCRAVDDQWPEEAVAPLYNYLVDVPGFDMSMVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHKLE
GLDSEALEITKYKGAFPYMNYLDEVGAYYTERIAVQATQVRSLDFSIIMMSQDQERIESQTSATNVATLM
QNAGTKVAGKIVSDDKTAKTISNAAGQEARANMVSLQRQDGLIGTSWIDGNQINIQMENKINVQDLITLQ
PGENYTVFQGDPVPGASFYIEPEDKTCGEPLIINRYITINPPGLKQLRKLVPRTAQRRLPTPENVSKIIG
VLTAKPSRRKKKSGGEPWKVIDTFQQRLANRQEAHNLMTEYDMDHSGREKSLWEEALEVIRSTTMAERTI
RYITLNRPEPEPSEPGDEENSQPQEVILKNLTLPAPVIRPARTGNEPPQQISPHIPLRPEMEWPE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 8261..40724
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| OCT50_RS22170 (OCT50_22165) | 3626..4372 | + | 747 | WP_289897194 | TcpQ domain-containing protein | - |
| OCT50_RS22175 (OCT50_22170) | 4384..4809 | + | 426 | WP_231070154 | type IV pilus biogenesis protein PilM | - |
| OCT50_RS22180 (OCT50_22175) | 4822..6498 | + | 1677 | WP_172693695 | PilN family type IVB pilus formation outer membrane protein | - |
| OCT50_RS22185 (OCT50_22180) | 6500..7783 | + | 1284 | WP_289897195 | type 4b pilus protein PilO2 | - |
| OCT50_RS22190 (OCT50_22185) | 7767..8258 | + | 492 | WP_289897196 | type IV pilus biogenesis protein PilP | - |
| OCT50_RS22195 (OCT50_22190) | 8261..9820 | + | 1560 | WP_289897197 | GspE/PulE family protein | virB11 |
| OCT50_RS22200 (OCT50_22195) | 9813..10907 | + | 1095 | WP_289897198 | type II secretion system F family protein | - |
| OCT50_RS22205 (OCT50_22200) | 10922..11536 | + | 615 | WP_084833199 | type 4 pilus major pilin | - |
| OCT50_RS22210 (OCT50_22205) | 11546..12103 | + | 558 | WP_151587085 | lytic transglycosylase domain-containing protein | virB1 |
| OCT50_RS22215 (OCT50_22210) | 12088..12762 | + | 675 | WP_289897199 | prepilin peptidase | - |
| OCT50_RS22220 (OCT50_22215) | 12759..14066 | + | 1308 | WP_289897200 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| OCT50_RS22225 (OCT50_22220) | 14173..14658 | + | 486 | WP_289897201 | hypothetical protein | - |
| OCT50_RS22230 (OCT50_22225) | 14789..15625 | + | 837 | WP_289897202 | hypothetical protein | traE |
| OCT50_RS22235 (OCT50_22230) | 15727..16932 | + | 1206 | WP_289897203 | conjugal transfer protein TraF | - |
| OCT50_RS22240 (OCT50_22235) | 17097..17555 | + | 459 | WP_172693685 | DotD/TraH family lipoprotein | - |
| OCT50_RS22245 (OCT50_22240) | 17552..18358 | + | 807 | WP_289897204 | type IV secretory system conjugative DNA transfer family protein | traI |
| OCT50_RS22250 (OCT50_22245) | 18373..19533 | + | 1161 | WP_289897205 | plasmid transfer ATPase TraJ | virB11 |
| OCT50_RS22255 (OCT50_22250) | 19530..19817 | + | 288 | WP_039031267 | IcmT/TraK family protein | traK |
| OCT50_RS22260 (OCT50_22255) | 19898..23509 | + | 3612 | WP_289897206 | LPD7 domain-containing protein | - |
| OCT50_RS22265 (OCT50_22260) | 23569..23877 | + | 309 | WP_289897215 | conjugal transfer protein | traL |
| OCT50_RS22270 (OCT50_22265) | 23883..24584 | + | 702 | WP_172693681 | DotI/IcmL/TraM family protein | traM |
| OCT50_RS22275 (OCT50_22270) | 24593..25582 | + | 990 | WP_172693680 | DotH/IcmK family type IV secretion protein | traN |
| OCT50_RS22280 (OCT50_22275) | 25582..26883 | + | 1302 | WP_289897207 | conjugal transfer protein TraO | traO |
| OCT50_RS22285 (OCT50_22280) | 26886..27671 | + | 786 | WP_289897208 | conjugal transfer protein TraP | traP |
| OCT50_RS22290 (OCT50_22285) | 27671..28195 | + | 525 | WP_289897209 | conjugal transfer protein TraQ | traQ |
| OCT50_RS22295 (OCT50_22290) | 28228..28632 | + | 405 | WP_289897210 | DUF6750 family protein | traR |
| OCT50_RS22300 (OCT50_22295) | 28676..29533 | + | 858 | WP_289897211 | conjugal transfer protein TraT | traT |
| OCT50_RS22305 (OCT50_22300) | 29604..32657 | + | 3054 | WP_289897212 | ATP-binding protein | traU |
| OCT50_RS22310 (OCT50_22305) | 32654..33226 | + | 573 | WP_241281501 | conjugal transfer protein TraV | traV |
| OCT50_RS22315 (OCT50_22310) | 33242..34441 | + | 1200 | WP_289897216 | conjugal transfer protein TraW | traW |
| OCT50_RS22320 (OCT50_22315) | 34438..34983 | + | 546 | WP_208690799 | conjugal transfer protein TraX | - |
| OCT50_RS22325 (OCT50_22320) | 35047..37206 | + | 2160 | WP_241281503 | DotA/TraY family protein | traY |
| OCT50_RS22330 (OCT50_22325) | 37266..37898 | + | 633 | WP_172693670 | plasmid IncI1-type surface exclusion protein ExcA | - |
| OCT50_RS22335 (OCT50_22330) | 38223..39434 | + | 1212 | WP_231070079 | conjugal transfer protein TrbA | trbA |
| OCT50_RS22340 (OCT50_22335) | 39486..40724 | + | 1239 | WP_241281504 | hypothetical protein | trbB |
| OCT50_RS22345 (OCT50_22340) | 40728..43025 | + | 2298 | WP_241281505 | F-type conjugative transfer protein TrbC | - |
| OCT50_RS22350 (OCT50_22345) | 43142..45031 | - | 1890 | WP_231070075 | DUF262 domain-containing protein | - |
Host bacterium
| ID | 13931 | GenBank | NZ_CP106960 |
| Plasmid name | 17YN198|p1 | Incompatibility group | - |
| Plasmid size | 115001 bp | Coordinate of oriT [Strand] | 48189..48272 [+] |
| Host baterium | Leclercia adecarboxylata strain 17YN198 |
Cargo genes
| Drug resistance gene | tet(A), aph(3')-Ia, qnrB19, sul1, qacE, ant(3'')-Ia, dfrA1 |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |