Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113426 |
| Name | oriT_pESI-CTX-M-1 |
| Organism | Salmonella enterica subsp. enterica serovar Infantis isolate 12037823-11 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_OW849779 (234137..234225 [+], 89 nt) |
| oriT length | 89 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 89 nt
>oriT_pESI-CTX-M-1
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTC
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file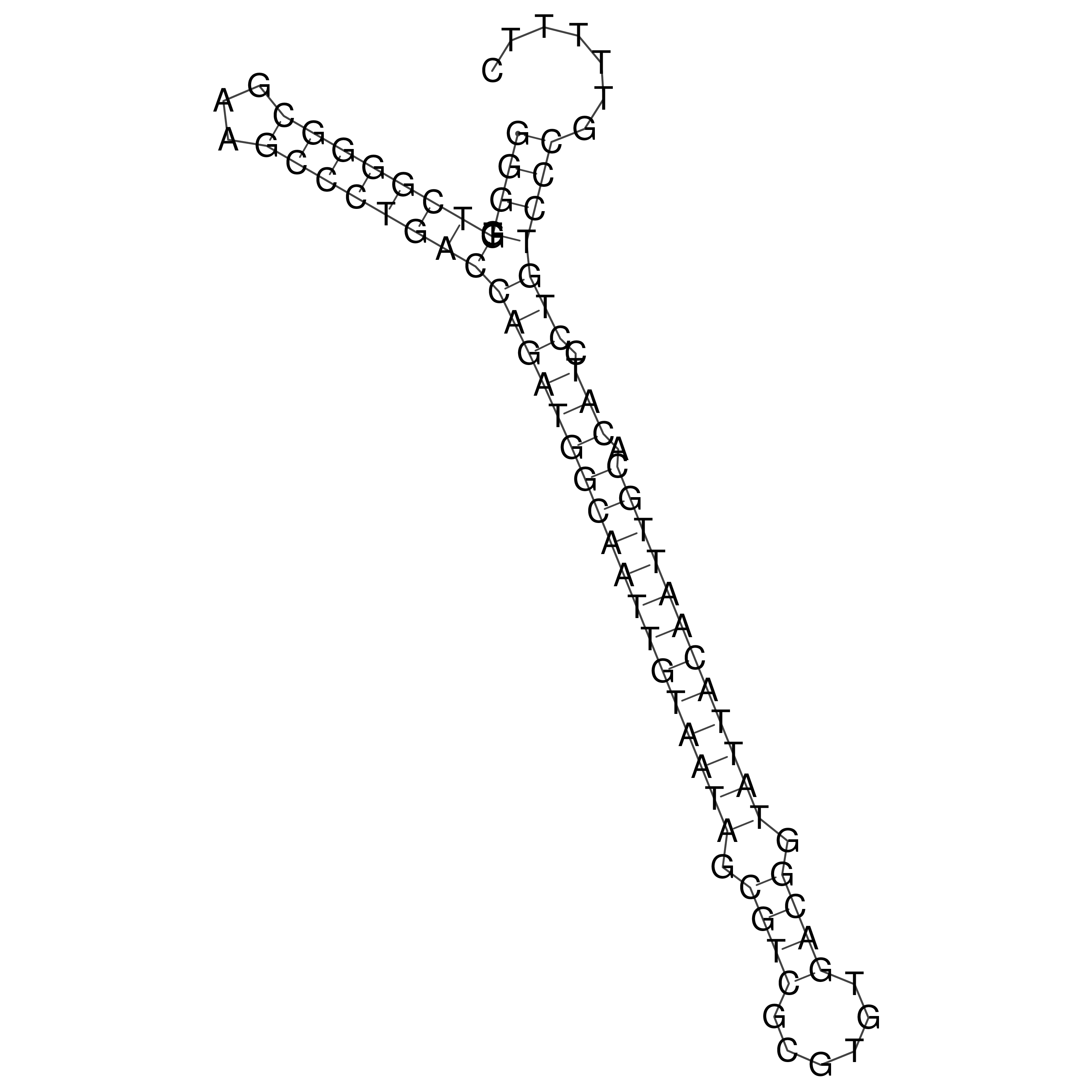
Relaxase
| ID | 8604 | GenBank | WP_001299212 |
| Name | nikB_NH452_RS01135_pESI-CTX-M-1 |
UniProt ID | A0A5T4AK12 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103956.40 Da Isoelectric Point: 7.2642
>WP_001299212.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWINFYGVTCFHNCTSLETAAADMEYIALQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVSDWLSLHRRLAEDGLYLSQMDGK
YLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWINFYGVTCFHNCTSLETAAADMEYIALQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVSDWLSLHRRLAEDGLYLSQMDGK
YLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5T4AK12 |
Auxiliary protein
| ID | 4997 | GenBank | WP_001283947 |
| Name | WP_001283947_pESI-CTX-M-1 |
UniProt ID | A0A142CMC2 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12613.57 Da Isoelectric Point: 10.7463
>WP_001283947.1 MULTISPECIES: IncI1-type relaxosome accessory protein NikA [Enterobacteriaceae]
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A142CMC2 |
T4CP
| ID | 9938 | GenBank | WP_001289276 |
| Name | trbC_NH452_RS01130_pESI-CTX-M-1 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86941.11 Da Isoelectric Point: 6.7713
>WP_001289276.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKTNRPDWDGMY
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKTNRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 192617..228759
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NH452_RS00925 | 187679..188746 | + | 1068 | WP_001360287 | type IV pilus biogenesis lipoprotein PilL | - |
| NH452_RS00930 | 188746..189183 | + | 438 | WP_000539807 | type IV pilus biogenesis protein PilM | - |
| NH452_RS00935 | 189197..190879 | + | 1683 | WP_000748143 | PilN family type IVB pilus formation outer membrane protein | - |
| NH452_RS00940 | 190872..192167 | + | 1296 | WP_001545757 | type 4b pilus protein PilO2 | - |
| NH452_RS00945 | 192154..192606 | + | 453 | WP_001247331 | type IV pilus biogenesis protein PilP | - |
| NH452_RS00950 | 192617..194170 | + | 1554 | WP_000362206 | GspE/PulE family protein | virB11 |
| NH452_RS00955 | 194183..195268 | + | 1086 | WP_001208802 | type II secretion system F family protein | - |
| NH452_RS00960 | 195285..195899 | + | 615 | WP_000908226 | type 4 pilus major pilin | - |
| NH452_RS00965 | 195909..196469 | + | 561 | WP_000014005 | lytic transglycosylase domain-containing protein | virB1 |
| NH452_RS00970 | 196454..197110 | + | 657 | WP_001193549 | A24 family peptidase | - |
| NH452_RS00975 | 197110..198402 | + | 1293 | WP_001350014 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| NH452_RS00980 | 198458..198583 | - | 126 | Protein_203 | RNA-guided endonuclease TnpB family protein | - |
| NH452_RS00985 | 198791..199615 | + | 825 | WP_001238942 | conjugal transfer protein TraE | traE |
| NH452_RS00990 | 199701..200903 | + | 1203 | WP_000976348 | conjugal transfer protein TraF | - |
| NH452_RS00995 | 200963..201547 | + | 585 | WP_000977522 | histidine phosphatase family protein | - |
| NH452_RS01000 | 201942..202400 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| NH452_RS01005 | 202397..203215 | + | 819 | WP_000646097 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| NH452_RS01010 | 203212..204360 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| NH452_RS01015 | 204357..204647 | + | 291 | WP_001299214 | hypothetical protein | traK |
| NH452_RS01020 | 204662..205213 | + | 552 | WP_000014584 | phospholipase D family protein | - |
| NH452_RS01025 | 205303..209070 | + | 3768 | WP_001141542 | LPD7 domain-containing protein | - |
| NH452_RS01030 | 209088..209435 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| NH452_RS01035 | 209432..210124 | + | 693 | WP_000138552 | DotI/IcmL family type IV secretion protein | traM |
| NH452_RS01040 | 210135..211118 | + | 984 | WP_001191878 | IncI1-type conjugal transfer protein TraN | traN |
| NH452_RS01045 | 211121..212410 | + | 1290 | WP_001271997 | conjugal transfer protein TraO | traO |
| NH452_RS01050 | 212410..213114 | + | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| NH452_RS01055 | 213114..213641 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| NH452_RS01060 | 213692..214096 | + | 405 | WP_000086961 | IncI1-type conjugal transfer protein TraR | traR |
| NH452_RS01065 | 214160..214348 | + | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| NH452_RS01070 | 214332..215132 | + | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| NH452_RS01075 | 215222..218266 | + | 3045 | WP_001024780 | IncI1-type conjugal transfer protein TraU | traU |
| NH452_RS01080 | 218266..218880 | + | 615 | WP_000337400 | IncI1-type conjugal transfer protein TraV | traV |
| NH452_RS01085 | 218847..220049 | + | 1203 | WP_001189156 | IncI1-type conjugal transfer protein TraW | traW |
| NH452_RS01090 | 220078..220662 | + | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| NH452_RS01095 | 220759..222927 | + | 2169 | WP_000698357 | DotA/TraY family protein | traY |
| NH452_RS01100 | 223001..223651 | + | 651 | WP_001178506 | plasmid IncI1-type surface exclusion protein ExcA | - |
| NH452_RS01105 | 223723..223932 | - | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| NH452_RS01110 | 224216..224500 | + | 285 | WP_215198320 | hypothetical protein | - |
| NH452_RS01500 | 224565..224660 | - | 96 | WP_001303310 | DinQ-like type I toxin DqlB | - |
| NH452_RS01115 | 225292..226200 | + | 909 | WP_000055084 | hypothetical protein | - |
| NH452_RS01120 | 226462..227670 | + | 1209 | WP_000121273 | IncI1-type conjugal transfer protein TrbA | trbA |
| NH452_RS01125 | 227689..228759 | + | 1071 | WP_000151577 | IncI1-type conjugal transfer protein TrbB | trbB |
| NH452_RS01130 | 228752..231043 | + | 2292 | WP_001289276 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 13861 | GenBank | NZ_OW849779 |
| Plasmid name | pESI-CTX-M-1 | Incompatibility group | IncFIB |
| Plasmid size | 290187 bp | Coordinate of oriT [Strand] | 234137..234225 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Infantis isolate 12037823-11 |
Cargo genes
| Drug resistance gene | blaCTX-M-1, dfrA1, ant(3'')-Ia, qacE, sul1, tet(A), dfrA14, aph(3')-Ia |
| Virulence gene | faeC, faeD, faeE, faeF, faeH, faeI, fyuA, ybtE, ybtT, ybtU, irp1, irp2, ybtA, ybtP, ybtQ, ybtX, ybtS |
| Metal resistance gene | merE, merD, merA, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |