Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113418 |
| Name | oriT_pIncI1-ST26 |
| Organism | Salmonella enterica subsp. enterica serovar Heidelberg isolate FT2-21BPWGent |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP128391 (41409..41495 [-], 87 nt) |
| oriT length | 87 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 87 nt
>oriT_pIncI1-ST26
GGGGTGTCGGGGCGAAGCCCTGACCAGGAGGCAATTGTATAATCGCGCGTGCGCGGTTATACAATTGCACATCCTGTCCCGTTTTTC
GGGGTGTCGGGGCGAAGCCCTGACCAGGAGGCAATTGTATAATCGCGCGTGCGCGGTTATACAATTGCACATCCTGTCCCGTTTTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file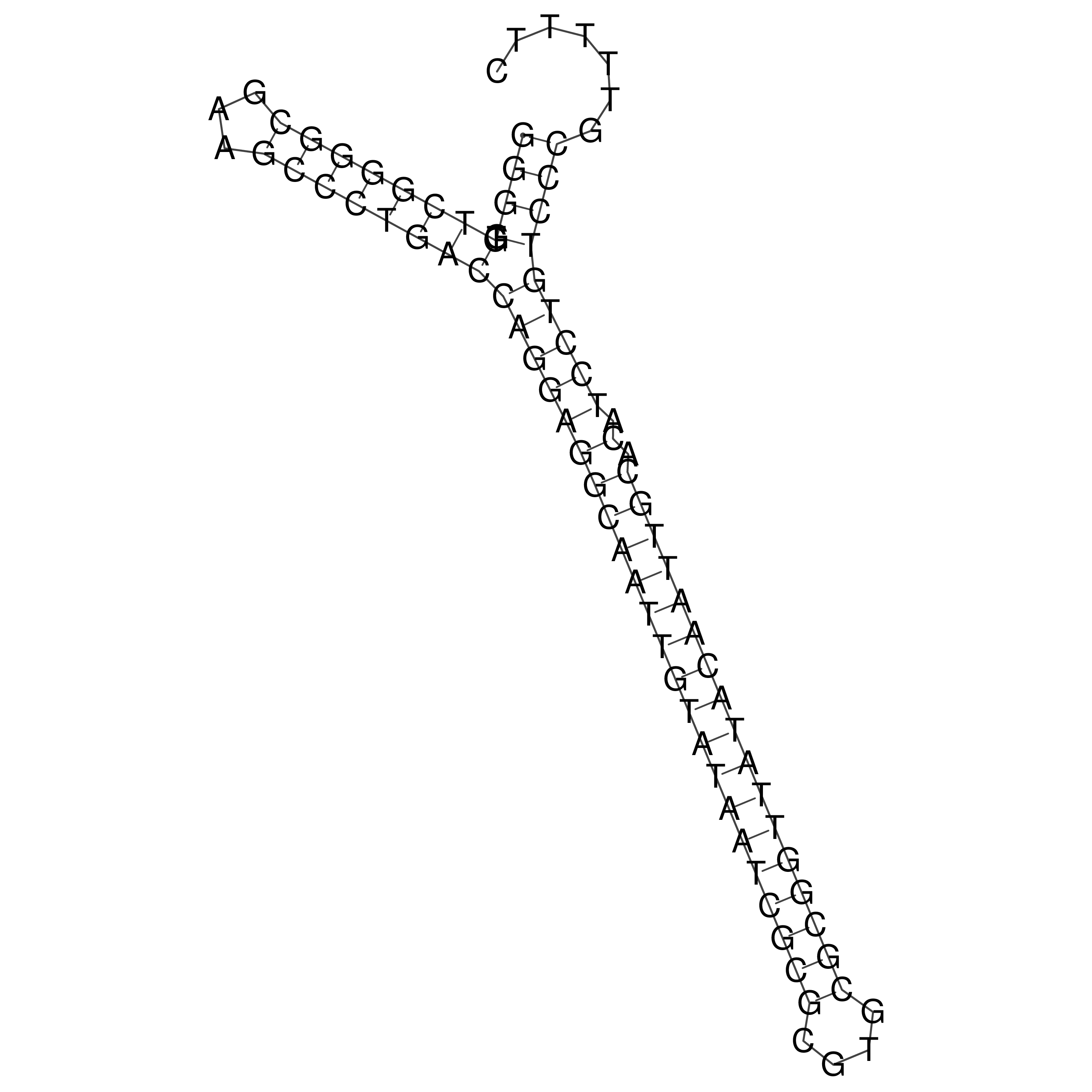
Relaxase
| ID | 8598 | GenBank | WP_001354015 |
| Name | nikB_QS243_RS23590_pIncI1-ST26 |
UniProt ID | E8XLT1 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104018.47 Da Isoelectric Point: 7.3526
>WP_001354015.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCLAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPENRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCLAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPENRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | E8XLT1 |
Auxiliary protein
| ID | 2376 | GenBank | WP_001281051 |
| Name | WP_001281051_pIncI1-ST26 |
UniProt ID | A0A3U8I0C6 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12646.44 Da Isoelectric Point: 10.4030
>WP_001281051.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSDKRQRSGSERRQKTILRAVRFSPDEDETICKNAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
MSDKRQRSGSERRQKTILRAVRFSPDEDETICKNAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3U8I0C6 |
T4CP
| ID | 9935 | GenBank | WP_001289276 |
| Name | trbC_QS243_RS23595_pIncI1-ST26 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86941.11 Da Isoelectric Point: 6.7713
>WP_001289276.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKTNRPDWDGMY
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKTNRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 46874..76304
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QS243_RS23595 (QS243_23595) | 44590..46881 | - | 2292 | WP_001289276 | F-type conjugative transfer protein TrbC | - |
| QS243_RS23600 (QS243_23600) | 46874..47944 | - | 1071 | WP_000151583 | IncI1-type conjugal transfer protein TrbB | trbB |
| QS243_RS23605 (QS243_23605) | 47963..49171 | - | 1209 | WP_000121274 | IncI1-type conjugal transfer protein TrbA | trbA |
| QS243_RS23610 (QS243_23610) | 49463..49615 | + | 153 | WP_001331364 | Hok/Gef family protein | - |
| QS243_RS24225 | 49687..49878 | - | 192 | WP_174715448 | hypothetical protein | - |
| QS243_RS23615 (QS243_23615) | 50439..50534 | + | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| QS243_RS23620 (QS243_23620) | 50599..50775 | - | 177 | WP_001054904 | hypothetical protein | - |
| QS243_RS23625 (QS243_23625) | 51167..51376 | + | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| QS243_RS23630 (QS243_23630) | 51448..52110 | - | 663 | WP_000653334 | plasmid IncI1-type surface exclusion protein ExcA | - |
| QS243_RS23635 (QS243_23635) | 52175..54337 | - | 2163 | WP_000698351 | DotA/TraY family protein | traY |
| QS243_RS23640 (QS243_23640) | 54434..55018 | - | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| QS243_RS23645 (QS243_23645) | 55047..56249 | - | 1203 | WP_001189160 | IncI1-type conjugal transfer protein TraW | traW |
| QS243_RS23650 (QS243_23650) | 56216..56830 | - | 615 | WP_000337398 | IncI1-type conjugal transfer protein TraV | traV |
| QS243_RS23655 (QS243_23655) | 56830..59874 | - | 3045 | WP_001024780 | IncI1-type conjugal transfer protein TraU | traU |
| QS243_RS23660 (QS243_23660) | 59964..60764 | - | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| QS243_RS23665 (QS243_23665) | 60748..60936 | - | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| QS243_RS23670 (QS243_23670) | 61000..61404 | - | 405 | WP_000086957 | IncI1-type conjugal transfer protein TraR | traR |
| QS243_RS23675 (QS243_23675) | 61455..61982 | - | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| QS243_RS23680 (QS243_23680) | 61982..62686 | - | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| QS243_RS23685 (QS243_23685) | 62686..63975 | - | 1290 | WP_001272005 | conjugal transfer protein TraO | traO |
| QS243_RS23690 (QS243_23690) | 63978..64961 | - | 984 | WP_001191877 | IncI1-type conjugal transfer protein TraN | traN |
| QS243_RS23695 (QS243_23695) | 64972..65664 | - | 693 | WP_000138548 | DotI/IcmL family type IV secretion protein | traM |
| QS243_RS23700 (QS243_23700) | 65661..66008 | - | 348 | WP_001055900 | conjugal transfer protein | traL |
| QS243_RS23705 (QS243_23705) | 66026..69793 | - | 3768 | WP_001141529 | LPD7 domain-containing protein | - |
| QS243_RS23710 (QS243_23710) | 69883..70434 | - | 552 | WP_000014583 | phospholipase D family protein | - |
| QS243_RS23715 (QS243_23715) | 70449..70739 | - | 291 | WP_001299214 | hypothetical protein | traK |
| QS243_RS23720 (QS243_23720) | 70736..71884 | - | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| QS243_RS23725 (QS243_23725) | 71881..72699 | - | 819 | WP_000646098 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| QS243_RS23730 (QS243_23730) | 72696..73154 | - | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| QS243_RS23735 (QS243_23735) | 73549..74133 | - | 585 | WP_000977522 | histidine phosphatase family protein | - |
| QS243_RS23740 (QS243_23740) | 74193..75395 | - | 1203 | WP_000976351 | conjugal transfer protein TraF | - |
| QS243_RS23745 (QS243_23745) | 75480..76304 | - | 825 | WP_001238939 | conjugal transfer protein TraE | traE |
| QS243_RS23750 (QS243_23750) | 76455..77609 | - | 1155 | WP_001139957 | tyrosine-type recombinase/integrase | - |
Host bacterium
| ID | 13853 | GenBank | NZ_CP128391 |
| Plasmid name | pIncI1-ST26 | Incompatibility group | - |
| Plasmid size | 77659 bp | Coordinate of oriT [Strand] | 41409..41495 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Heidelberg isolate FT2-21BPWGent |
Cargo genes
| Drug resistance gene | aac(3)-VIa, ant(3'')-Ia |
| Virulence gene | - |
| Metal resistance gene | merR1, merT, merP, merA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |