Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113098 |
| Name | oriT_10290|pla1 |
| Organism | Escherichia sp. 10290 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP025742 (121243..121366 [-], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 92..99, 113..120 (ATAATGTA..TACATTAT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTATTTGAATCATTAACTTATGTCTTAAATAATGTATTTTAATTTATTTTACATTATAAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file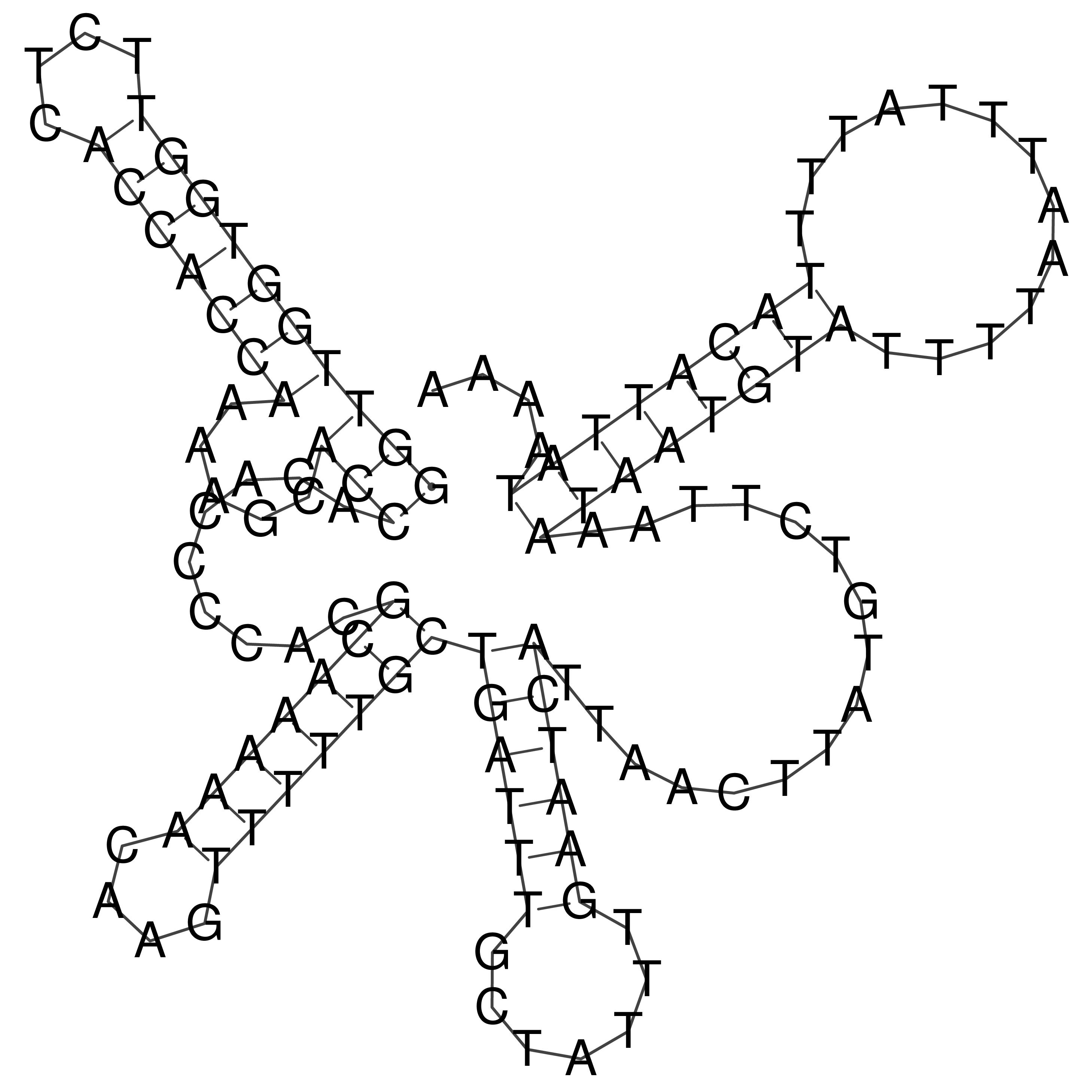
Relaxase
| ID | 8407 | GenBank | WP_000986971 |
| Name | traI_QUE74_RS27845_10290|pla1 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192129.86 Da Isoelectric Point: 5.8508
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGKLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMPGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIRAAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAESRMQTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTTGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELAMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLISDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKIPVAEGERLRVTGKIPGLRISGGDRLQVASVSEDAMTVVVPGRAEPASLPVGYSPFTALKL
ESGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVVESKNLAFSQVDLLTEAKSFAAEGTSFTDLGREINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETIDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWENNPDAIVLVDNVYHRIAGISRDDGLITLQDTDGNSRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVMAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIALGRKYPQPYVALPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSEDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAEEKPDQP
DDKTEQAVRDIAGQERDRAVTSEREAVLPESVLREPQRVQDAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 4814 | GenBank | WP_001254381 |
| Name | WP_001254381_10290|pla1 |
UniProt ID | _ |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9037.10 Da Isoelectric Point: 9.9851
MRRRNAKAGTSRTISVYLDEDTNNRLIRAKDRSGRSKTIEVQIRLRDHLKRYPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4815 | GenBank | WP_001151555 |
| Name | WP_001151555_10290|pla1 |
UniProt ID | _ |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14461.36 Da Isoelectric Point: 4.5221
MAKVQAYVSDEIVYKINEIVERRRAEGAKSTDVSFSSISTMLLELGLRVYEAQMERKESAFNQAEFNKVL
LECAIKTQSTVAKILGIESLSPHVSGNPKFEYANMVEDIRDKVSSEMERFFPENDEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 9684 | GenBank | WP_000009375 |
| Name | traD_QUE74_RS27860_10290|pla1 |
UniProt ID | _ |
| Length | 714 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 714 a.a. Molecular weight: 81137.72 Da Isoelectric Point: 5.1669
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKLSWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATIVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDLNPEMENRLSAVLAAREAEGRQMASLFEPEVVSGEDVTQAEQPQQPVSSVINDK
KSDAGVSVPAGGIAQELKMKPEEEMEQQLPPGISESGEVVDMAVYEAWQREQNPDIQQQMQRREEVNINV
HRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 9685 | GenBank | WP_001064271 |
| Name | traC_QUE74_RS27975_10290|pla1 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99434.22 Da Isoelectric Point: 5.9665
MNNPLEAVTQAVNSLVTALKLPDESSKANEILGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANETIVEALDHMLRTKLPRGIPLCIHLMSSQLVGERIKYGLREFSWSGEQAERFNAITRAYYMKAAE
TLFPLPEGLNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGAYITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLVKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRQEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 92912..121938
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QUE74_RS27850 (EsCdI10290_05518) | 92278..92505 | + | 228 | WP_000450526 | toxin-antitoxin system antitoxin VapB | - |
| QUE74_RS27855 (EsCdI10290_05519) | 92505..92903 | + | 399 | WP_000911316 | type II toxin-antitoxin system VapC family toxin | - |
| QUE74_RS27860 (EsCdI10290_05520) | 92912..95056 | - | 2145 | WP_000009375 | type IV conjugative transfer system coupling protein TraD | virb4 |
| QUE74_RS27865 (EsCdI10290_05521) | 95112..95842 | - | 731 | Protein_112 | hypothetical protein | - |
| QUE74_RS27870 (EsCdI10290_05522) | 96077..96556 | - | 480 | WP_001355324 | hypothetical protein | - |
| QUE74_RS27875 (EsCdI10290_05523) | 96760..97491 | - | 732 | WP_001364068 | conjugal transfer complement resistance protein TraT | - |
| QUE74_RS27880 (EsCdI10290_05524) | 97523..98026 | - | 504 | WP_000605506 | hypothetical protein | - |
| QUE74_RS27885 (EsCdI10290_05525) | 98042..100885 | - | 2844 | WP_001006995 | conjugal transfer mating-pair stabilization protein TraG | traG |
| QUE74_RS27890 (EsCdI10290_05526) | 100882..102255 | - | 1374 | WP_001137366 | conjugal transfer pilus assembly protein TraH | traH |
| QUE74_RS27895 (EsCdI10290_05527) | 102242..102634 | - | 393 | WP_000138366 | F-type conjugal transfer protein TrbF | - |
| QUE74_RS27900 (EsCdI10290_05528) | 102588..102902 | - | 315 | WP_001244895 | P-type conjugative transfer protein TrbJ | - |
| QUE74_RS27905 (EsCdI10290_05529) | 102892..103428 | - | 537 | WP_000059854 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| QUE74_RS27910 | 103415..103699 | - | 285 | WP_000624195 | type-F conjugative transfer system pilin chaperone TraQ | - |
| QUE74_RS27915 | 103780..104115 | + | 336 | WP_000415768 | hypothetical protein | - |
| QUE74_RS27920 | 104096..104401 | - | 306 | WP_236922840 | conjugal transfer protein TrbA | - |
| QUE74_RS27925 (EsCdI10290_05531) | 104446..105189 | - | 744 | WP_001030399 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| QUE74_RS27930 (EsCdI10290_05532) | 105182..105439 | - | 258 | WP_000864356 | conjugal transfer protein TrbE | - |
| QUE74_RS27935 | 105453..107357 | - | 1905 | WP_000662057 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| QUE74_RS27940 | 107385..107603 | + | 219 | WP_001364024 | hypothetical protein | - |
| QUE74_RS27945 | 107592..107972 | - | 381 | WP_000009645 | hypothetical protein | - |
| QUE74_RS27950 (EsCdI10290_05534) | 107969..108607 | - | 639 | WP_000087541 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| QUE74_RS29505 | 108634..109153 | - | 520 | Protein_130 | hypothetical protein | - |
| QUE74_RS27960 (EsCdI10290_05536) | 109429..110421 | - | 993 | WP_001355328 | conjugal transfer pilus assembly protein TraU | traU |
| QUE74_RS27965 (EsCdI10290_05537) | 110418..111050 | - | 633 | WP_000650119 | type-F conjugative transfer system protein TraW | traW |
| QUE74_RS27970 (EsCdI10290_05538) | 111047..111433 | - | 387 | WP_000214089 | type-F conjugative transfer system protein TrbI | - |
| QUE74_RS27975 (EsCdI10290_05539) | 111430..114057 | - | 2628 | WP_001064271 | type IV secretion system protein TraC | virb4 |
| QUE74_RS27980 (EsCdI10290_05540) | 114217..114438 | - | 222 | WP_001278688 | conjugal transfer protein TraR | - |
| QUE74_RS27985 (EsCdI10290_05541) | 114573..115088 | - | 516 | WP_000802746 | type IV conjugative transfer system lipoprotein TraV | traV |
| QUE74_RS27990 (EsCdI10290_05542) | 115085..115405 | - | 321 | WP_001057269 | DUF2689 domain-containing protein | - |
| QUE74_RS27995 (EsCdI10290_05543) | 115392..115979 | - | 588 | WP_000002892 | conjugal transfer pilus-stabilizing protein TraP | - |
| QUE74_RS28000 (EsCdI10290_05544) | 115969..117399 | - | 1431 | WP_000146619 | F-type conjugal transfer pilus assembly protein TraB | traB |
| QUE74_RS28005 (EsCdI10290_05545) | 117399..118127 | - | 729 | WP_001355329 | type-F conjugative transfer system secretin TraK | traK |
| QUE74_RS28010 (EsCdI10290_05546) | 118114..118680 | - | 567 | WP_000399757 | type IV conjugative transfer system protein TraE | traE |
| QUE74_RS28015 (EsCdI10290_05547) | 118702..119013 | - | 312 | WP_000012121 | type IV conjugative transfer system protein TraL | traL |
| QUE74_RS28020 (EsCdI10290_05548) | 119028..119387 | - | 360 | WP_000340277 | type IV conjugative transfer system pilin TraA | - |
| QUE74_RS28025 (EsCdI10290_05549) | 119421..119648 | - | 228 | WP_001254381 | conjugal transfer relaxosome protein TraY | - |
| QUE74_RS28030 (EsCdI10290_05550) | 119742..120428 | - | 687 | WP_000332479 | PAS domain-containing protein | - |
| QUE74_RS28035 (EsCdI10290_05551) | 120622..121005 | - | 384 | WP_001151555 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| QUE74_RS28040 (EsCdI10290_05552) | 121291..121938 | + | 648 | WP_000614283 | transglycosylase SLT domain-containing protein | virB1 |
| QUE74_RS28045 (EsCdI10290_05553) | 122235..123056 | - | 822 | Protein_148 | DUF932 domain-containing protein | - |
| QUE74_RS28050 (EsCdI10290_05554) | 123175..123462 | - | 288 | WP_000107535 | hypothetical protein | - |
| QUE74_RS28055 (EsCdI10290_05555) | 123713..124717 | + | 1005 | WP_024166184 | IS110 family transposase | - |
| QUE74_RS28060 | 124811..125071 | - | 261 | WP_000547943 | hypothetical protein | - |
| QUE74_RS28065 | 125141..125313 | + | 173 | Protein_152 | hypothetical protein | - |
| QUE74_RS28070 | 125738..125863 | - | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| QUE74_RS28075 (EsCdI10290_05556) | 125805..125954 | - | 150 | Protein_154 | plasmid maintenance protein Mok | - |
| QUE74_RS28080 | 125976..126164 | + | 189 | WP_001355331 | hypothetical protein | - |
| QUE74_RS28085 (EsCdI10290_05557) | 126109..126895 | - | 787 | Protein_156 | plasmid SOS inhibition protein A | - |
Host bacterium
| ID | 13533 | GenBank | NZ_AP025742 |
| Plasmid name | 10290|pla1 | Incompatibility group | IncFIB |
| Plasmid size | 224339 bp | Coordinate of oriT [Strand] | 121243..121366 [-] |
| Host baterium | Escherichia sp. 10290 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hlyC, faeJ, faeI, faeH, faeF, faeE, faeD, faeC, espP |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF11 |