Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113085 |
| Name | oriT_pNARUSE |
| Organism | Bacillus subtilis subsp. natto strain NARUSE |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP028060 (28908..29269 [+], 362 nt) |
| oriT length | 362 nt |
| IRs (inverted repeats) | 188..193, 198..203 (CACCAT..ATGGTG) 171..178, 180..187 (AAACCGCA..TGCGGTTT) 116..122, 130..136 (ACATAAA..TTTATGT) 84..89, 101..106 (ATTTAG..CTAAAT) 84..89, 92..97 (ATTTAG..CTAAAT) 62..67, 75..80 (CATAAA..TTTATG) 48..54, 65..71 (TTATTTT..AAAATAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 362 nt
>oriT_pNARUSE
AAAGAGCAATCTCGTCATCGAAGACTAAATTTCTGTATGGAAAACAGTTATTTTTCGTGTGCATAAAATAAAGATTTATGTGCATTTAGTTCTAAATCACCTAAATAATGGTTGAACATAAATGTTTTTTTTATGTGCATTCAGCAATAAATCTGGTACCACGAAAAAACAAACCGCACTGCGGTTTCACCATGCAAATGGTGCCAGTTTCCCCTTATGCTCTTTCCTTTTCCACCCCCTCCTTCCTTCGGAATCGGGGGCCGGCTTTTTGCTGCCGCAAAAAGCCTTGGAAAAAGAAACACTTATTTGAACAGATCGTTGCAAAGTAATGTGCAGAAATCGATGAAAAGGAGCGTTAACAA
AAAGAGCAATCTCGTCATCGAAGACTAAATTTCTGTATGGAAAACAGTTATTTTTCGTGTGCATAAAATAAAGATTTATGTGCATTTAGTTCTAAATCACCTAAATAATGGTTGAACATAAATGTTTTTTTTATGTGCATTCAGCAATAAATCTGGTACCACGAAAAAACAAACCGCACTGCGGTTTCACCATGCAAATGGTGCCAGTTTCCCCTTATGCTCTTTCCTTTTCCACCCCCTCCTTCCTTCGGAATCGGGGGCCGGCTTTTTGCTGCCGCAAAAAGCCTTGGAAAAAGAAACACTTATTTGAACAGATCGTTGCAAAGTAATGTGCAGAAATCGATGAAAAGGAGCGTTAACAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file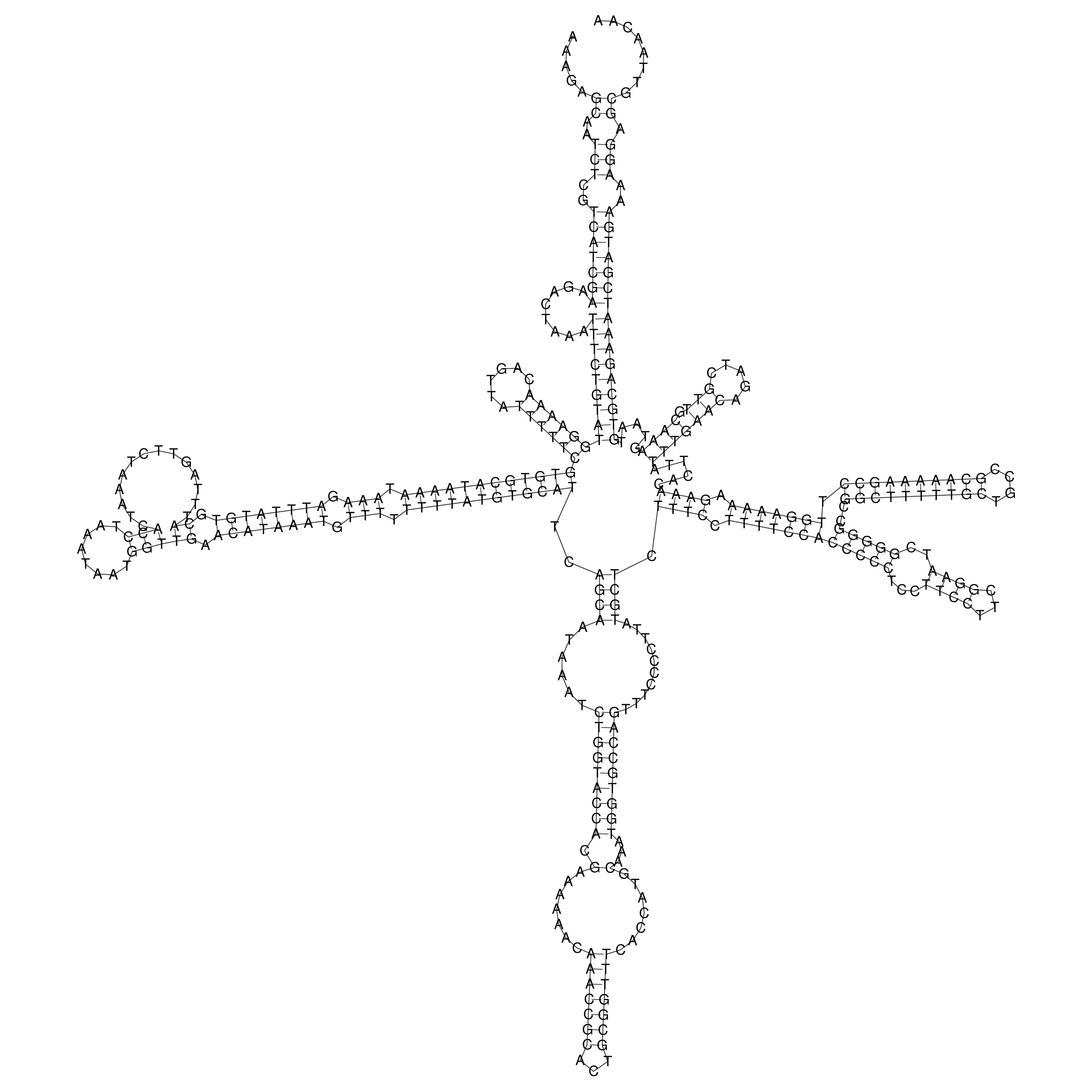
Relaxase
| ID | 8398 | GenBank | WP_013603221 |
| Name | mobP2_QUE82_RS22265_pNARUSE |
UniProt ID | E9RJ23 |
| Length | 410 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 410 a.a. Molecular weight: 48596.08 Da Isoelectric Point: 10.0206
>WP_013603221.1 MULTISPECIES: MobP2 family relaxase [Bacillaceae]
MDSPGVVLVSKYVSGKSTKFSKYVNYINRDEAVRTEKFQTYNVNKLDGYNQYMGNPEKSSGIFTQHKDSL
SPVEKNQLKEIFRQAQKNDSVMWQDVISFDNKWLEERGIYNSQTGWVNEGAIQNSIRKGMEVLLREEQLE
QSGVWSAAIHYNTDNIHVHIALVEPNPTKEYGVFTNKKTGEVYQARRGNRKLKTLDKMKSKVANTLMDRD
KELSKISQLIHDRIAPKGLKFQPRLDTYMTKMYNHIYENLPEDMRLWKYNNNALNNIRPEIDSVITMYIQ
KYHPEDYKELDQSLKEEMEFRKSVYGDGPKQVERYKEYRKNKHKELYTKLGNSMLKEMAEIRRRESQSKQ
MNRSGTYAPSSVSNRQWDTKGRRIKRSDINKIKHALDKDYQSMKNMRKYQQMQYEMEQSR
MDSPGVVLVSKYVSGKSTKFSKYVNYINRDEAVRTEKFQTYNVNKLDGYNQYMGNPEKSSGIFTQHKDSL
SPVEKNQLKEIFRQAQKNDSVMWQDVISFDNKWLEERGIYNSQTGWVNEGAIQNSIRKGMEVLLREEQLE
QSGVWSAAIHYNTDNIHVHIALVEPNPTKEYGVFTNKKTGEVYQARRGNRKLKTLDKMKSKVANTLMDRD
KELSKISQLIHDRIAPKGLKFQPRLDTYMTKMYNHIYENLPEDMRLWKYNNNALNNIRPEIDSVITMYIQ
KYHPEDYKELDQSLKEEMEFRKSVYGDGPKQVERYKEYRKNKHKELYTKLGNSMLKEMAEIRRRESQSKQ
MNRSGTYAPSSVSNRQWDTKGRRIKRSDINKIKHALDKDYQSMKNMRKYQQMQYEMEQSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | E9RJ23 |
T4CP
| ID | 9672 | GenBank | WP_013603210 |
| Name | t4cp2_QUE82_RS22215_pNARUSE |
UniProt ID | _ |
| Length | 786 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 786 a.a. Molecular weight: 90624.17 Da Isoelectric Point: 4.9504
>WP_013603210.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Bacillus]
MHKEFKQWKKVFADKYFLIAFTLFCYVAVTCITNFFIHLLKQIPVILSSFKNFDQSQLSSPFSSMTWKWF
FEFDWSMGIVYGVVYVLASIFILRQVYRFRIAFRDINKQTKGTARWTEIMEIQETYKAVKDDDNEYEGSA
GMPIVHFNDHLYVDTNSTHTLVVASTQSGKTETYSYPYLDVIMRAKEKDSVVITDIKGDMLKNTRAEFEK
YGYEVMCFNLLNPYWGMAYNPLELVKQAYLKKDYSKAQMLCNTLSYSLFHNDKSHADPMWENASIALVNA
LILAVCDLCIKNGQPENITMYTLTVMLNELGSNPDEDGYTRLDHFFGNLPPSHPAKLQYGTIQFSQGITR
SGIYTGTMAKLKNYTYDTIARMTARNDLNIEDLAYGEKPVALFIVYPDWDDSNYTIISTFLSQVNAVLSE
KATLSKESTLPRKVRFLFEEVANIPPIEGLNRSLAVGLSRGMLYTLVIQNVSQLRDVYGDDMAVAIMGNL
GNQIYIMSDEWEDAEKFSEKLGVTTVISADRQGDLMDVHKSYSEREEERPLMLPDELRRLKKGEWVVLRT
KKREDLKRNRVVPYPIFASLDNGTNMLHRYEYLMHRFDNKIALGDLGFRGEHETLDLESLLIEFEFNEPV
KEERKSKPGKGKKQKAKKAPGEPVSSEFPLPEEPIGFTESSEGMDSLFASIPVENMNEHGFYEDPPMNES
LHEDDLEASEEDIIMNSLEEVYETSTPISDAIKPDHYVYIKLLAHKQLSESEYAYFDSLKTVEELRAFFR
APEKKEIYEQIKTHIE
MHKEFKQWKKVFADKYFLIAFTLFCYVAVTCITNFFIHLLKQIPVILSSFKNFDQSQLSSPFSSMTWKWF
FEFDWSMGIVYGVVYVLASIFILRQVYRFRIAFRDINKQTKGTARWTEIMEIQETYKAVKDDDNEYEGSA
GMPIVHFNDHLYVDTNSTHTLVVASTQSGKTETYSYPYLDVIMRAKEKDSVVITDIKGDMLKNTRAEFEK
YGYEVMCFNLLNPYWGMAYNPLELVKQAYLKKDYSKAQMLCNTLSYSLFHNDKSHADPMWENASIALVNA
LILAVCDLCIKNGQPENITMYTLTVMLNELGSNPDEDGYTRLDHFFGNLPPSHPAKLQYGTIQFSQGITR
SGIYTGTMAKLKNYTYDTIARMTARNDLNIEDLAYGEKPVALFIVYPDWDDSNYTIISTFLSQVNAVLSE
KATLSKESTLPRKVRFLFEEVANIPPIEGLNRSLAVGLSRGMLYTLVIQNVSQLRDVYGDDMAVAIMGNL
GNQIYIMSDEWEDAEKFSEKLGVTTVISADRQGDLMDVHKSYSEREEERPLMLPDELRRLKKGEWVVLRT
KKREDLKRNRVVPYPIFASLDNGTNMLHRYEYLMHRFDNKIALGDLGFRGEHETLDLESLLIEFEFNEPV
KEERKSKPGKGKKQKAKKAPGEPVSSEFPLPEEPIGFTESSEGMDSLFASIPVENMNEHGFYEDPPMNES
LHEDDLEASEEDIIMNSLEEVYETSTPISDAIKPDHYVYIKLLAHKQLSESEYAYFDSLKTVEELRAFFR
APEKKEIYEQIKTHIE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 13520 | GenBank | NZ_AP028060 |
| Plasmid name | pNARUSE | Incompatibility group | - |
| Plasmid size | 62770 bp | Coordinate of oriT [Strand] | 28908..29269 [+] |
| Host baterium | Bacillus subtilis subsp. natto strain NARUSE |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |