Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 112896 |
| Name | oriT_pPM400 |
| Organism | Bacillus badius strain NBPM-293 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP082364 (40720..40761 [-], 42 nt) |
| oriT length | 42 nt |
| IRs (inverted repeats) | 3..8, 16..21 (CCCCAC..GTGGGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 42 nt
>oriT_pPM400
AACCCCACACGTTAAGTGGGGGCATTTTTCCCTTATGCTCTT
AACCCCACACGTTAAGTGGGGGCATTTTTCCCTTATGCTCTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file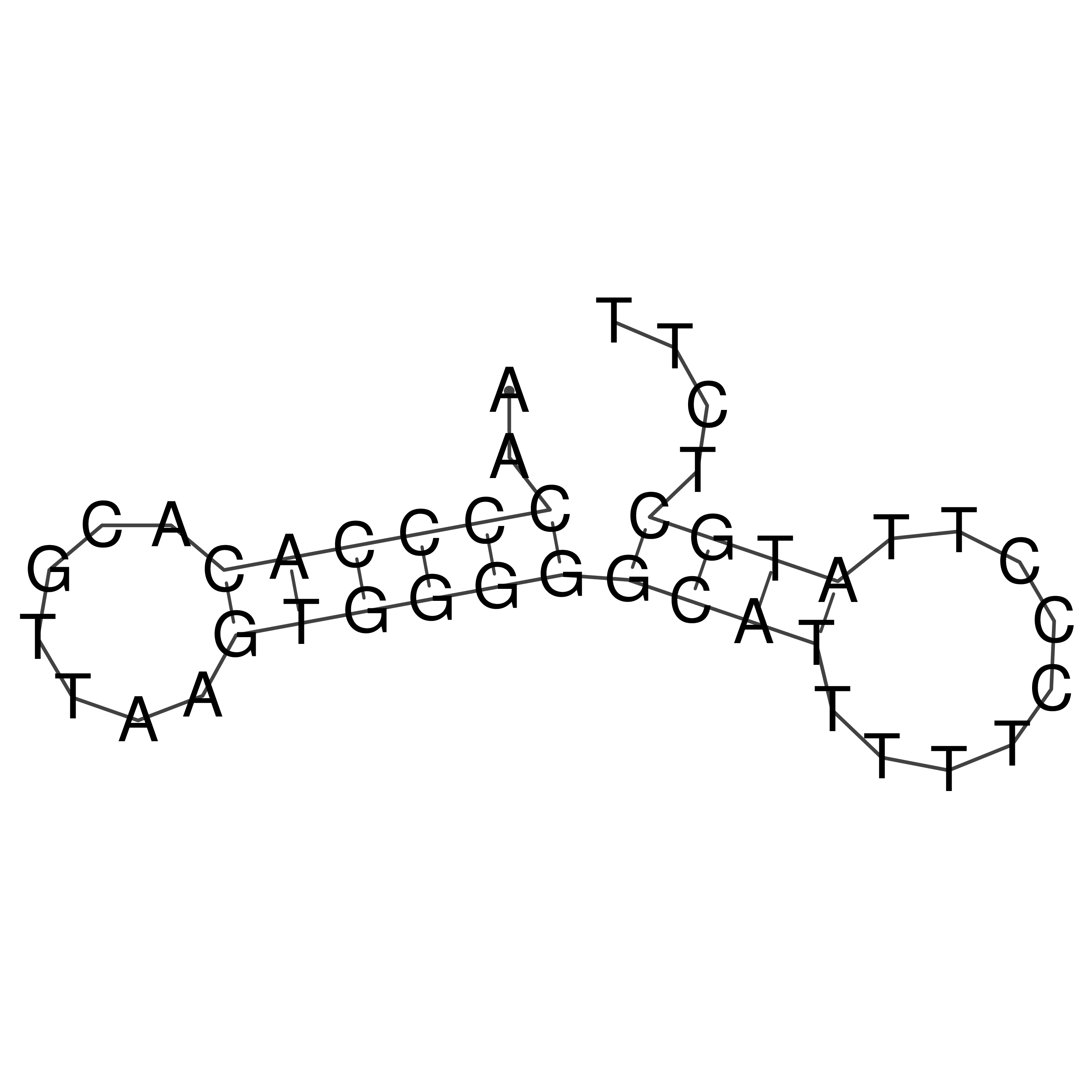
Relaxase
| ID | 8299 | GenBank | WP_041098556 |
| Name | mobP2_K7T73_RS20125_pPM400 |
UniProt ID | _ |
| Length | 409 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 409 a.a. Molecular weight: 48956.34 Da Isoelectric Point: 9.8878
>WP_041098556.1 MobP2 family relaxase [Bacillus badius]
MSRSPGVVLKSKFVMPSASEYNNYVEYMDRDDVKREVQVNRHSQAVNDFQVYHSYINYMGDEEKQGSLFT
SDIDELNSKQKRELKKIFHLAQQNGSPLWQDVISFDNQWLQKQALYDSKTGALDEEKIRNVVREAIKEML
KAENMEDTAVWSAAIHYNTDNIHVHVATVEPFPTREKKRIFDKETNEWTEQYRAKRKQGTLDKMKSRVAN
TILDRTKERNRINDLIRGSVSTKKSVDLATYRKTKRLFQKAIKKLPEDRKKWQYGYQAINDARPYIDEIV
EVYLNTYHKKEMRELHKLLDDEVKVMKELYGEGSQYQKYKETKLNDLQKRLGNAVLVEIRAYDKRVRQME
AQTFYAKNHRSYTQYKWRRSQSLDSAIRNLQFRLRKSFHDYQKDRNVEEFDRMLDGYER
MSRSPGVVLKSKFVMPSASEYNNYVEYMDRDDVKREVQVNRHSQAVNDFQVYHSYINYMGDEEKQGSLFT
SDIDELNSKQKRELKKIFHLAQQNGSPLWQDVISFDNQWLQKQALYDSKTGALDEEKIRNVVREAIKEML
KAENMEDTAVWSAAIHYNTDNIHVHVATVEPFPTREKKRIFDKETNEWTEQYRAKRKQGTLDKMKSRVAN
TILDRTKERNRINDLIRGSVSTKKSVDLATYRKTKRLFQKAIKKLPEDRKKWQYGYQAINDARPYIDEIV
EVYLNTYHKKEMRELHKLLDDEVKVMKELYGEGSQYQKYKETKLNDLQKRLGNAVLVEIRAYDKRVRQME
AQTFYAKNHRSYTQYKWRRSQSLDSAIRNLQFRLRKSFHDYQKDRNVEEFDRMLDGYER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 9621 | GenBank | WP_223664252 |
| Name | t4cp2_K7T73_RS20200_pPM400 |
UniProt ID | _ |
| Length | 740 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 740 a.a. Molecular weight: 84077.05 Da Isoelectric Point: 5.3346
>WP_223664252.1 VirD4-like conjugal transfer protein, CD1115 family [Bacillus badius]
MILVSGFLFVSFFLANFLLAFLQELLMALKQFSENPETVDFSLEWGSFFVFQKTLLPFYIGFYIIALIGV
VKFIYNVRMNYQSLNEGQHGTSEFETVKELKKQYKVIPASEKEYEGHGGVIVAGLQEKHRPYRLLIDEGP
IHSMVIGITRSGKGETFVVPMIDVISRAKEKASMVVNDPKGELAAASYETLKERGYEVYIFNLIQQQMSM
GFNPLEIVVDAWKKGKTSFAQQYANSVAFSLYNDPNAKDPYWSNSAKSLVTAIILALTEDAVATGKEEKV
NMYSVANFLSTLGSDNDDQGNNALDLFFKSRAENNPARMMYATSNFAAGNTRASIFSTAMDKLQIFTLEP
NAKLTSYNSLPLTRIGFGDKPVAIFMATPDYDESNHVLASIFVSQLYRINAEKATMSPSGKMKRQVHFLL
DEFGNMPTVDGMSSMVTVGAGRGFRFHLIVQAYSQLKTKYGDEADTIIGNCSNQIYILTKDQKTAEQFSS
LVGKKTITDVSRSGKLLSTDKSHSESTKERPLLMPDELMRLKEGESVVVRANKRQDNKRRKIEPKPIYNR
GVTAHKFRWEYLADDFDTSKSLMELPILSNDYHDMNLREMVFTTKTESDQFVPIVSLIGKEAFLQFKKEV
RKLLIITGEDLPLNNIDEWTAIQLFSYLTFQTECDPGLFTMMINKCLATYLPESVTNDWIERMKLVREKQ
QEQKNEVNKLEDRLEEENEENYSDCGDEEDIKQQLKNEIM
MILVSGFLFVSFFLANFLLAFLQELLMALKQFSENPETVDFSLEWGSFFVFQKTLLPFYIGFYIIALIGV
VKFIYNVRMNYQSLNEGQHGTSEFETVKELKKQYKVIPASEKEYEGHGGVIVAGLQEKHRPYRLLIDEGP
IHSMVIGITRSGKGETFVVPMIDVISRAKEKASMVVNDPKGELAAASYETLKERGYEVYIFNLIQQQMSM
GFNPLEIVVDAWKKGKTSFAQQYANSVAFSLYNDPNAKDPYWSNSAKSLVTAIILALTEDAVATGKEEKV
NMYSVANFLSTLGSDNDDQGNNALDLFFKSRAENNPARMMYATSNFAAGNTRASIFSTAMDKLQIFTLEP
NAKLTSYNSLPLTRIGFGDKPVAIFMATPDYDESNHVLASIFVSQLYRINAEKATMSPSGKMKRQVHFLL
DEFGNMPTVDGMSSMVTVGAGRGFRFHLIVQAYSQLKTKYGDEADTIIGNCSNQIYILTKDQKTAEQFSS
LVGKKTITDVSRSGKLLSTDKSHSESTKERPLLMPDELMRLKEGESVVVRANKRQDNKRRKIEPKPIYNR
GVTAHKFRWEYLADDFDTSKSLMELPILSNDYHDMNLREMVFTTKTESDQFVPIVSLIGKEAFLQFKKEV
RKLLIITGEDLPLNNIDEWTAIQLFSYLTFQTECDPGLFTMMINKCLATYLPESVTNDWIERMKLVREKQ
QEQKNEVNKLEDRLEEENEENYSDCGDEEDIKQQLKNEIM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 41893..58095
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| K7T73_RS20120 (K7T73_20085) | 37760..38515 | - | 756 | WP_223664153 | Fic family protein | - |
| K7T73_RS20125 (K7T73_20090) | 38600..39829 | - | 1230 | WP_041098556 | MobP2 family relaxase | - |
| K7T73_RS20130 (K7T73_20095) | 39826..40269 | - | 444 | WP_041098553 | hypothetical protein | - |
| K7T73_RS20135 (K7T73_20100) | 40244..40585 | - | 342 | WP_041098551 | ribbon-helix-helix protein, CopG family | - |
| K7T73_RS20140 (K7T73_20105) | 40845..41840 | - | 996 | WP_052475181 | hypothetical protein | - |
| K7T73_RS20145 (K7T73_20110) | 41893..42954 | - | 1062 | WP_052475180 | NlpC/P60 family protein | orf14 |
| K7T73_RS20150 (K7T73_20115) | 42957..43556 | - | 600 | WP_041098549 | DUF6556 family protein | - |
| K7T73_RS20155 (K7T73_20120) | 43558..45513 | - | 1956 | WP_223664155 | VirB4 family type IV secretion system protein | virb4 |
| K7T73_RS20160 (K7T73_20125) | 45524..46192 | - | 669 | WP_041114328 | hypothetical protein | - |
| K7T73_RS20165 (K7T73_20130) | 46173..46529 | - | 357 | WP_041098543 | DUF5592 family protein | traL |
| K7T73_RS20170 (K7T73_20135) | 46526..48850 | - | 2325 | WP_223664157 | pLS20_p028 family conjugation system transmembrane protein | virB6 |
| K7T73_RS20175 (K7T73_20140) | 48885..49460 | - | 576 | WP_041098539 | hypothetical protein | - |
| K7T73_RS20180 (K7T73_20145) | 49486..50229 | - | 744 | WP_052475179 | type II secretion system F family protein | - |
| K7T73_RS20185 (K7T73_20150) | 50222..51343 | - | 1122 | WP_041098535 | ATPase, T2SS/T4P/T4SS family | virB11 |
| K7T73_RS20190 (K7T73_20155) | 51336..51605 | - | 270 | WP_223664159 | YdbC family protein | - |
| K7T73_RS20195 (K7T73_20160) | 51633..52334 | - | 702 | WP_041098533 | type II secretion system F family protein | - |
| K7T73_RS20200 (K7T73_20165) | 52353..54575 | - | 2223 | WP_223664252 | VirD4-like conjugal transfer protein, CD1115 family | - |
| K7T73_RS20205 (K7T73_20170) | 54656..55012 | - | 357 | WP_041098529 | hypothetical protein | - |
| K7T73_RS20210 (K7T73_20175) | 55024..55452 | - | 429 | WP_041098527 | hypothetical protein | - |
| K7T73_RS20215 (K7T73_20180) | 55459..56862 | - | 1404 | WP_223664161 | toprim domain-containing protein | - |
| K7T73_RS20220 (K7T73_20185) | 56825..57778 | - | 954 | WP_223664163 | hypothetical protein | - |
| K7T73_RS20225 (K7T73_20190) | 57862..58095 | - | 234 | WP_041098523 | pilin | virB2 |
| K7T73_RS20230 (K7T73_20195) | 58103..58354 | - | 252 | WP_082821652 | hypothetical protein | - |
| K7T73_RS20235 (K7T73_20200) | 58421..58588 | - | 168 | WP_169713124 | hypothetical protein | - |
| K7T73_RS20240 (K7T73_20205) | 58998..59396 | - | 399 | WP_063384069 | hypothetical protein | - |
| K7T73_RS20245 (K7T73_20210) | 59414..60106 | - | 693 | WP_063384068 | hypothetical protein | - |
| K7T73_RS20250 (K7T73_20215) | 60367..62661 | - | 2295 | WP_223664170 | hypothetical protein | - |
Host bacterium
| ID | 13331 | GenBank | NZ_CP082364 |
| Plasmid name | pPM400 | Incompatibility group | - |
| Plasmid size | 395960 bp | Coordinate of oriT [Strand] | 40720..40761 [-] |
| Host baterium | Bacillus badius strain NBPM-293 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrID1 |