Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 112894 |
| Name | oriT_pUAMSEL3 |
| Organism | Enterococcus faecalis strain UAMS_EL56 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP076491 (18356..18405 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pUAMSEL3
AATATCGCAACATGCTAGCATGTTGCTCTGCTCGCAAAAAGAAAGCCTAC
AATATCGCAACATGCTAGCATGTTGCTCTGCTCGCAAAAAGAAAGCCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file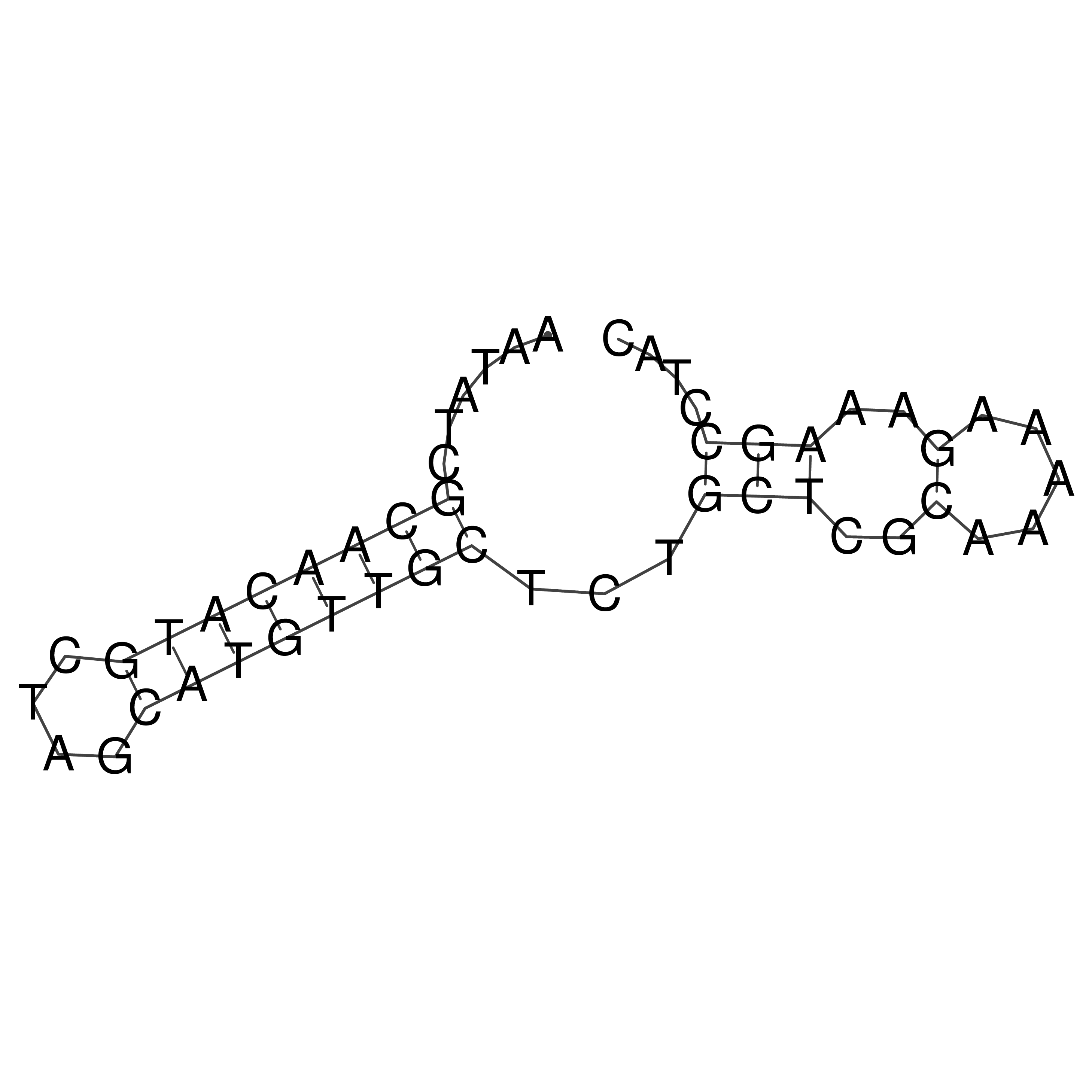
Relaxase
| ID | 8297 | GenBank | WP_002365873 |
| Name | Relaxase_KPU60_RS16055_pUAMSEL3 |
UniProt ID | _ |
| Length | 561 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65469.22 Da Isoelectric Point: 6.0174
>WP_002365873.1 relaxase/mobilization nuclease domain-containing protein [Enterococcus faecalis]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKRVFEQISDKIAAKEGAKIIEKSPKNS
HKKYTMWETESLYKEKIKSRLDFLLEQSSSINDFLEKAEALNLSVDFSKKWTTYRLLDEPQIKNTRSRSL
SKNDPTRYNYEKIVERLKENKSVLTLKEVVEKYVEKSEKTKNEFDYQLTIDEWQISHKTTRGYYVNVDFG
FGERGKVFIGAYKVDPLENGQYNIYVKRKDFFYLMNEKNADRNRYMTGETLIKQLRLYNGQTPLKKEPVM
RTIDELVNAINFLAANEIEDTRQLELLEEKLEAAFLEAEETLETLDEKMLELHQLSNLLLENELNGELEL
IQQKLKTLLPDATLAEFSYEDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKRVFEQISDKIAAKEGAKIIEKSPKNS
HKKYTMWETESLYKEKIKSRLDFLLEQSSSINDFLEKAEALNLSVDFSKKWTTYRLLDEPQIKNTRSRSL
SKNDPTRYNYEKIVERLKENKSVLTLKEVVEKYVEKSEKTKNEFDYQLTIDEWQISHKTTRGYYVNVDFG
FGERGKVFIGAYKVDPLENGQYNIYVKRKDFFYLMNEKNADRNRYMTGETLIKQLRLYNGQTPLKKEPVM
RTIDELVNAINFLAANEIEDTRQLELLEEKLEAAFLEAEETLETLDEKMLELHQLSNLLLENELNGELEL
IQQKLKTLLPDATLAEFSYEDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 4755 | GenBank | WP_002365872 |
| Name | WP_002365872_pUAMSEL3 |
UniProt ID | _ |
| Length | 118 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 118 a.a. Molecular weight: 13988.05 Da Isoelectric Point: 9.6548
>WP_002365872.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Enterococcus]
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVHSLNALVQSELNKLIKRKEQS
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVHSLNALVQSELNKLIKRKEQS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 9620 | GenBank | WP_242140990 |
| Name | t4cp2_KPU60_RS16035_pUAMSEL3 |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 609 a.a. Molecular weight: 70059.14 Da Isoelectric Point: 6.4636
>WP_242140990.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecalis]
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFHRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNVAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEVKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRALEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFHRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNVAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEVKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRALEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2785..15711
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KPU60_RS15965 (KPU60_15875) | 2401..2757 | + | 357 | WP_002365854 | pheromone response system RNA-binding regulator PrgU | - |
| KPU60_RS15970 (KPU60_15880) | 2785..3642 | + | 858 | WP_002401687 | pheromone response LPXTG-anchored adhesin PrgC | prgC |
| KPU60_RS15975 (KPU60_15885) | 3684..4583 | + | 900 | WP_025186515 | hypothetical protein | - |
| KPU60_RS15980 (KPU60_15890) | 4603..5037 | + | 435 | WP_002365857 | hypothetical protein | - |
| KPU60_RS15985 (KPU60_15895) | 5084..5311 | + | 228 | WP_002365858 | hypothetical protein | - |
| KPU60_RS15990 (KPU60_15900) | 5325..5621 | + | 297 | WP_002365860 | hypothetical protein | - |
| KPU60_RS15995 (KPU60_15905) | 5636..6436 | + | 801 | WP_002365861 | PrgH | prgHb |
| KPU60_RS16000 (KPU60_15910) | 6438..6791 | + | 354 | WP_002365862 | PrgI family mobile element protein | prgIc |
| KPU60_RS16005 (KPU60_15915) | 6745..9135 | + | 2391 | WP_002365863 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| KPU60_RS16010 (KPU60_15920) | 9147..11762 | + | 2616 | WP_002365864 | phage tail tip lysozyme | prgK |
| KPU60_RS16015 (KPU60_15925) | 11786..12412 | + | 627 | WP_242140985 | hypothetical protein | prgL |
| KPU60_RS16020 (KPU60_15930) | 12399..12644 | + | 246 | WP_002382804 | hypothetical protein | - |
| KPU60_RS16025 (KPU60_15935) | 12628..13236 | + | 609 | WP_242140987 | PcfA protein | - |
| KPU60_RS16030 (KPU60_15940) | 13397..13882 | + | 486 | WP_016616031 | PcfB family protein | gbs1365 |
| KPU60_RS16035 (KPU60_15945) | 13882..15711 | + | 1830 | WP_242140990 | VirD4-like conjugal transfer protein, CD1115 family | virb4 |
| KPU60_RS16040 (KPU60_15950) | 15762..17921 | + | 2160 | WP_002365871 | ArdC-like ssDNA-binding domain-containing protein | - |
| KPU60_RS16045 (KPU60_15955) | 17954..18226 | + | 273 | WP_002362409 | LtrD | - |
| KPU60_RS16050 (KPU60_15960) | 18472..18828 | + | 357 | WP_002365872 | plasmid mobilization relaxosome protein MobC | - |
| KPU60_RS16055 (KPU60_15965) | 18829..20514 | + | 1686 | WP_002365873 | relaxase/mobilization nuclease domain-containing protein | - |
Host bacterium
| ID | 13329 | GenBank | NZ_CP076491 |
| Plasmid name | pUAMSEL3 | Incompatibility group | - |
| Plasmid size | 53193 bp | Coordinate of oriT [Strand] | 18356..18405 [+] |
| Host baterium | Enterococcus faecalis strain UAMS_EL56 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | EF0149 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |