Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 112362 |
| Name | oriT_pPlaYM7902C |
| Organism | Pseudomonas amygdali pv. lachrymans strain YM7902 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP127048 (39235..39414 [+], 180 nt) |
| oriT length | 180 nt |
| IRs (inverted repeats) | 80..87, 91..98 (CAAAGGGG..CCCCTTTG) 42..47, 56..61 (AAAAAG..CTTTTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 180 nt
>oriT_pPlaYM7902C
ATCTCGAATTCGCTAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTAGCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCAGAGCGGGAAAGCGTTGTCGTTGTCGTTGTTGGTTGTCGTCATTCAGGTGCCCTCAGGAGGTGATGTTGTGGC
ATCTCGAATTCGCTAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTAGCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCAGAGCGGGAAAGCGTTGTCGTTGTCGTTGTTGGTTGTCGTCATTCAGGTGCCCTCAGGAGGTGATGTTGTGGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file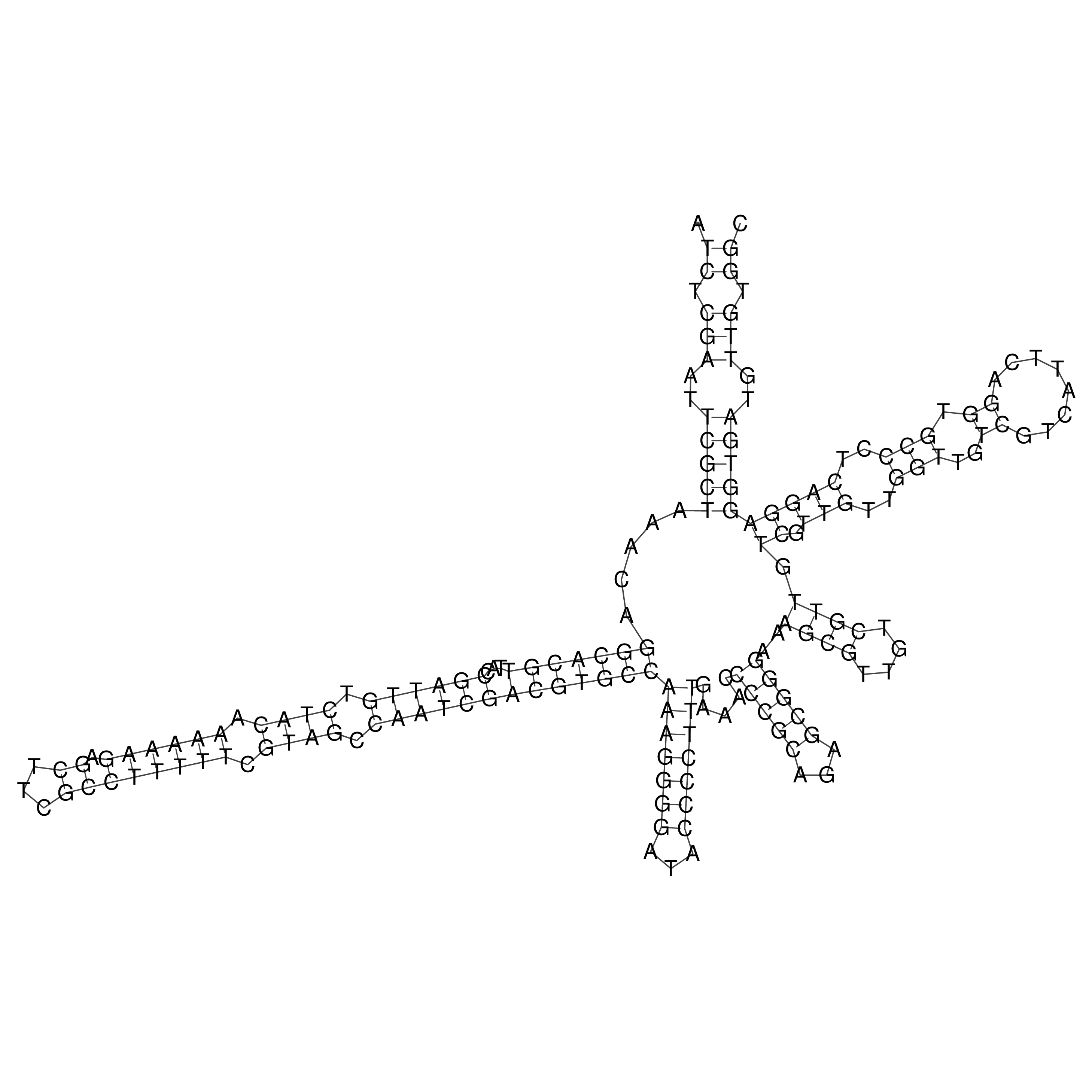
Relaxase
| ID | 7995 | GenBank | WP_052764764 |
| Name | Relaxase_QO021_RS31250_pPlaYM7902C |
UniProt ID | _ |
| Length | 416 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 416 a.a. Molecular weight: 47026.70 Da Isoelectric Point: 5.5334
>WP_052764764.1 relaxase/mobilization nuclease domain-containing protein [Pseudomonas amygdali]
MLIRVSGYNTGAQEYLEKGNKSGREFTRDELDHRLIIEGQLSLTRAIYESIPDYGQDRYLTFTLSFKEDT
VSPELLKSITTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDRKTGEVIDRKPHIHIIIPRINLLSGNEA
NPVDVYRNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQDLVKQVIER
GVTTRADFYALVAEHGETRIRNQGKDTEYISVKLPGDAKGTNLKDTIFQDDFIVRRELKKPPLEASVSAQ
ATPPRSKVIEGTLIEHGEAPYQHKDTNQLSYFVTLKPEGGKPRTVWGVGLEEAMTDANLKQGDQVRLEDL
GTWPVVVQVIAEDGTTSDKTVNRRAWSAQPVAPEREVAETTPKGQAIAAGTPELSSPDEEDGMSMD
MLIRVSGYNTGAQEYLEKGNKSGREFTRDELDHRLIIEGQLSLTRAIYESIPDYGQDRYLTFTLSFKEDT
VSPELLKSITTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDRKTGEVIDRKPHIHIIIPRINLLSGNEA
NPVDVYRNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQDLVKQVIER
GVTTRADFYALVAEHGETRIRNQGKDTEYISVKLPGDAKGTNLKDTIFQDDFIVRRELKKPPLEASVSAQ
ATPPRSKVIEGTLIEHGEAPYQHKDTNQLSYFVTLKPEGGKPRTVWGVGLEEAMTDANLKQGDQVRLEDL
GTWPVVVQVIAEDGTTSDKTVNRRAWSAQPVAPEREVAETTPKGQAIAAGTPELSSPDEEDGMSMD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 9196 | GenBank | WP_005746152 |
| Name | t4cp2_QO021_RS31205_pPlaYM7902C |
UniProt ID | _ |
| Length | 557 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 557 a.a. Molecular weight: 61711.38 Da Isoelectric Point: 6.4342
>WP_005746152.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Pseudomonas syringae group]
MARAKSKPISPDNPQERIEEHPAILIGKHPTKDTFLASYGQTFVMLAAPPGTGKTVGVVTPNLLSYPDSV
VVNDPKFENWRDTAGFRAAAGHKVYRFSPELLETHRWNPLSALSRDPLYRLGQIRTLAGVLFVSDNPKNQ
EWYNKAANVFAAILLYLMEMEGMKLNGMKLTLPQAYEVASLGTGLGVWAQQAIEQHSNGPNALSVETLRE
LNGVFEASKNKSSGWSTTVDILRGALSMYAEKTVAWAVSDTDIDFTKLRKEKISIYFCVTGNAIKKYGPL
MNLFFTQAIQQNTDVMPEQGGHCADGTLILKYQVLFVIDEIAVMGRIEIMEMAPALTRGAGLRYIIIFQG
KDQLRSERTYGREAGNGIMQAFHIEIVYGTGNVELAKEYSTRLGNTTVRVHNQSLNRQKHEIGARGQTDS
YSEQPRPLLLPQEVSALPYGEELIFVEATKTTPAANIRARKIFWYEEPVFKERAGMPTPDVPIGDASKID
ALTVPVRTVEAKVAVADTKPMQAEQRQRWNPKDKASEVAQAEAVKAQPVDVEPDPEPAQADDTLDTM
MARAKSKPISPDNPQERIEEHPAILIGKHPTKDTFLASYGQTFVMLAAPPGTGKTVGVVTPNLLSYPDSV
VVNDPKFENWRDTAGFRAAAGHKVYRFSPELLETHRWNPLSALSRDPLYRLGQIRTLAGVLFVSDNPKNQ
EWYNKAANVFAAILLYLMEMEGMKLNGMKLTLPQAYEVASLGTGLGVWAQQAIEQHSNGPNALSVETLRE
LNGVFEASKNKSSGWSTTVDILRGALSMYAEKTVAWAVSDTDIDFTKLRKEKISIYFCVTGNAIKKYGPL
MNLFFTQAIQQNTDVMPEQGGHCADGTLILKYQVLFVIDEIAVMGRIEIMEMAPALTRGAGLRYIIIFQG
KDQLRSERTYGREAGNGIMQAFHIEIVYGTGNVELAKEYSTRLGNTTVRVHNQSLNRQKHEIGARGQTDS
YSEQPRPLLLPQEVSALPYGEELIFVEATKTTPAANIRARKIFWYEEPVFKERAGMPTPDVPIGDASKID
ALTVPVRTVEAKVAVADTKPMQAEQRQRWNPKDKASEVAQAEAVKAQPVDVEPDPEPAQADDTLDTM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 17518..27872
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QO021_RS31085 (QO021_31080) | 13230..13511 | - | 282 | WP_002556029 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| QO021_RS31090 (QO021_31085) | 13511..13699 | - | 189 | WP_002556028 | hypothetical protein | - |
| QO021_RS31095 (QO021_31090) | 13888..14523 | - | 636 | WP_054068142 | recombinase family protein | - |
| QO021_RS31100 (QO021_31095) | 14838..15470 | + | 633 | WP_003381804 | ParA family protein | - |
| QO021_RS31105 (QO021_31100) | 15506..15760 | + | 255 | WP_003381801 | hypothetical protein | - |
| QO021_RS31110 (QO021_31105) | 16185..16850 | - | 666 | WP_004667059 | LexA family transcriptional regulator | - |
| QO021_RS31115 (QO021_31110) | 16938..17168 | + | 231 | WP_032704469 | Cro/CI family transcriptional regulator | - |
| QO021_RS31120 (QO021_31115) | 17518..18207 | + | 690 | WP_005746161 | lytic transglycosylase domain-containing protein | virB1 |
| QO021_RS31125 (QO021_31120) | 18204..18542 | + | 339 | WP_017279422 | hypothetical protein | - |
| QO021_RS31130 (QO021_31125) | 18555..19031 | + | 477 | WP_080544547 | VirB3 family type IV secretion system protein | virB3 |
| QO021_RS31135 (QO021_31130) | 18970..21444 | + | 2475 | WP_054068314 | VirB4 family type IV secretion system protein | virb4 |
| QO021_RS31140 (QO021_31135) | 21441..22127 | + | 687 | WP_004667979 | P-type DNA transfer protein VirB5 | - |
| QO021_RS31145 (QO021_31140) | 22160..22525 | + | 366 | WP_004643103 | hypothetical protein | - |
| QO021_RS31150 (QO021_31145) | 22536..23468 | + | 933 | WP_004667984 | type IV secretion system protein | virB6 |
| QO021_RS31155 (QO021_31150) | 23542..23808 | + | 267 | WP_005746157 | hypothetical protein | - |
| QO021_RS31160 (QO021_31155) | 23845..24633 | + | 789 | WP_004667986 | virB8 family protein | virB8 |
| QO021_RS31165 (QO021_31160) | 24623..25432 | + | 810 | WP_004667987 | P-type conjugative transfer protein VirB9 | virB9 |
| QO021_RS31170 (QO021_31165) | 25419..26807 | + | 1389 | WP_004667989 | TrbI/VirB10 family protein | virB10 |
| QO021_RS31175 (QO021_31170) | 26817..27872 | + | 1056 | WP_005746155 | P-type DNA transfer ATPase VirB11 | virB11 |
| QO021_RS31180 (QO021_31175) | 27886..28272 | + | 387 | WP_005746154 | hypothetical protein | - |
| QO021_RS31185 (QO021_31180) | 28257..28622 | + | 366 | WP_004667995 | hypothetical protein | - |
| QO021_RS31190 (QO021_31185) | 28669..28935 | + | 267 | WP_004644125 | hypothetical protein | - |
| QO021_RS31195 (QO021_31190) | 29060..31471 | + | 2412 | WP_044299713 | ATP-binding protein | - |
| QO021_RS31200 (QO021_31195) | 31587..31820 | + | 234 | WP_004666680 | hypothetical protein | - |
Host bacterium
| ID | 12797 | GenBank | NZ_CP127048 |
| Plasmid name | pPlaYM7902C | Incompatibility group | - |
| Plasmid size | 53818 bp | Coordinate of oriT [Strand] | 39235..39414 [+] |
| Host baterium | Pseudomonas amygdali pv. lachrymans strain YM7902 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |