Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 112018 |
| Name | oriT_pADAP |
| Organism | Serratia entomophila |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_002523 (133975..134057 [+], 83 nt) |
| oriT length | 83 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 83 nt
>oriT_pADAP
ACGGGACCAGATGTGTTTTGTAGCACCGCCTGCACGCAGTCGGTTCAACAAAACGCTGGGTCAGGGCAGAGCCCCGACACCCC
ACGGGACCAGATGTGTTTTGTAGCACCGCCTGCACGCAGTCGGTTCAACAAAACGCTGGGTCAGGGCAGAGCCCCGACACCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file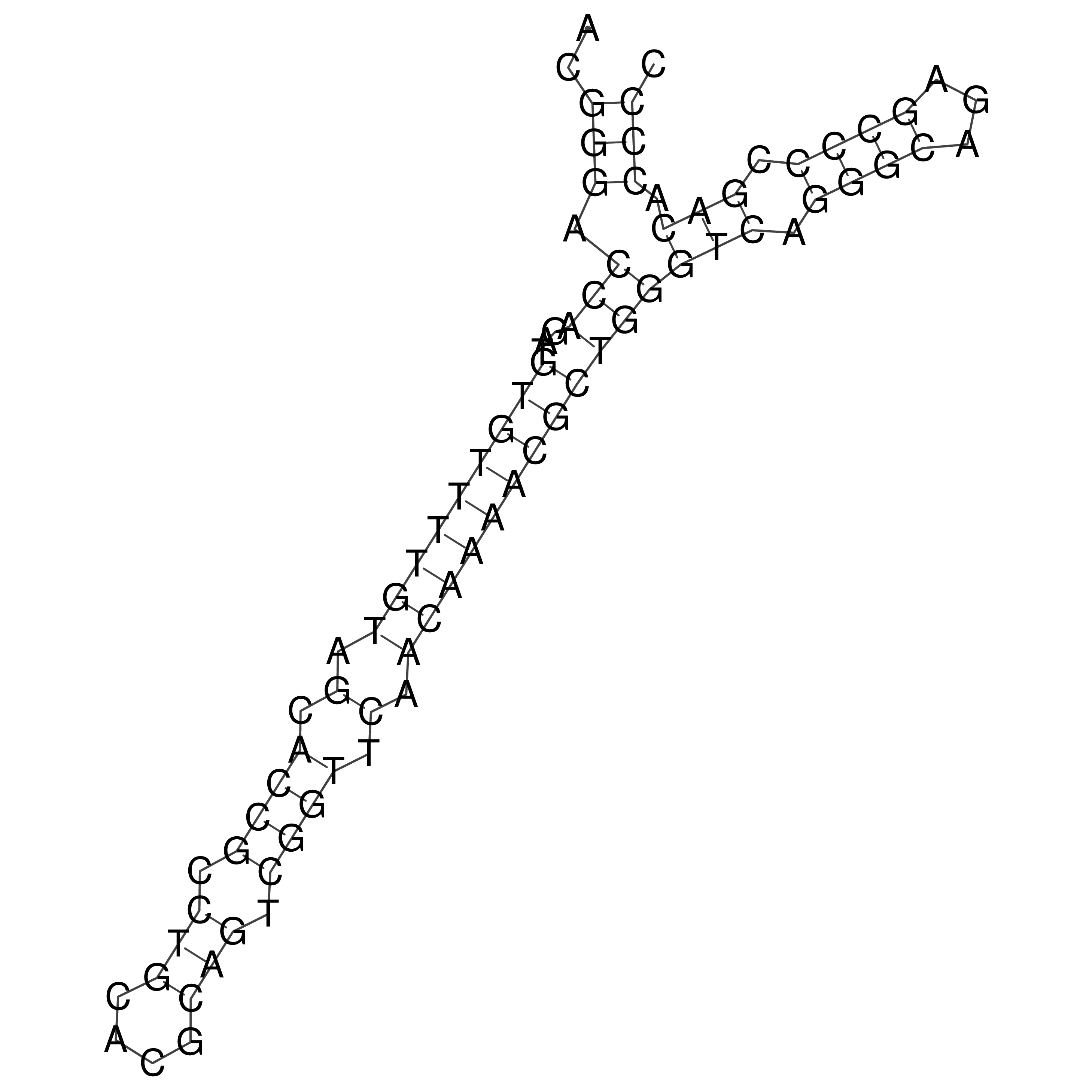
Relaxase
| ID | 7748 | GenBank | WP_010895859 |
| Name | Relaxase_HKJ47_RS00590_pADAP |
UniProt ID | A7M7H3 |
| Length | 651 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 651 a.a. Molecular weight: 73395.58 Da Isoelectric Point: 7.4747
>WP_010895859.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Serratia]
MIIRIPEKRRDGKSSFLQLVAYTVVRDEDKPDSPLEPEHPGWRRPRSKDAIFNHLVDYISRNGDADLQQI
MHADPNGHTQVMFDGVMCETNCFNIATASVEMNAVALQNTRCKDPVLHYFLSWPETDNPSTEHIFDSVRH
SLSALGMSEHQYVAAIHTDTNNIHCHVAANRIHPETYKAADDSFTRIRLQQAARELELKYNWTPTNGFFA
VNEQGEIVRSKREKNLTPSGAKTLEYYADVESLHTYAVTECGFKIDEAMADPNLAWKDIHCILVNAGLAL
KPKGKGLAIYSIDNPELPPVKASSVHPDLTLNCLEKDLGQFQLLENAGTYTKEHTTTAADAIVHEFRYEP
TLHARDLTARLERRLARADARADLKARYQAYKNAWKRPKLDAGAVKRRYQNESKRFAWQKARARVAIGDP
LLRKLTYHIIEVERMKAMAALRLTVKDERAAFKADPANRRLSYREWVEQQALSHDQAAIAQLRAFAYRMK
RTQRTAPLSTNSIVCAVADDTPAFALEGYATRVTRDGTVQYVRDGRVQLQDKGERIDVGDHRVNEGEHIA
GGMALAENKSGEHLKFEGEAAFVQQACSMVPWFNEGGDAPLPLSDPQQRTMAGYESTYQSASTGEGQRKP
SAVEQPEVEQNKPRSGHRPQQ
MIIRIPEKRRDGKSSFLQLVAYTVVRDEDKPDSPLEPEHPGWRRPRSKDAIFNHLVDYISRNGDADLQQI
MHADPNGHTQVMFDGVMCETNCFNIATASVEMNAVALQNTRCKDPVLHYFLSWPETDNPSTEHIFDSVRH
SLSALGMSEHQYVAAIHTDTNNIHCHVAANRIHPETYKAADDSFTRIRLQQAARELELKYNWTPTNGFFA
VNEQGEIVRSKREKNLTPSGAKTLEYYADVESLHTYAVTECGFKIDEAMADPNLAWKDIHCILVNAGLAL
KPKGKGLAIYSIDNPELPPVKASSVHPDLTLNCLEKDLGQFQLLENAGTYTKEHTTTAADAIVHEFRYEP
TLHARDLTARLERRLARADARADLKARYQAYKNAWKRPKLDAGAVKRRYQNESKRFAWQKARARVAIGDP
LLRKLTYHIIEVERMKAMAALRLTVKDERAAFKADPANRRLSYREWVEQQALSHDQAAIAQLRAFAYRMK
RTQRTAPLSTNSIVCAVADDTPAFALEGYATRVTRDGTVQYVRDGRVQLQDKGERIDVGDHRVNEGEHIA
GGMALAENKSGEHLKFEGEAAFVQQACSMVPWFNEGGDAPLPLSDPQQRTMAGYESTYQSASTGEGQRKP
SAVEQPEVEQNKPRSGHRPQQ
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A7M7H3 |
T4CP
| ID | 8861 | GenBank | WP_010895767 |
| Name | t4cp2_HKJ47_RS00135_pADAP |
UniProt ID | _ |
| Length | 577 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 577 a.a. Molecular weight: 64155.03 Da Isoelectric Point: 5.2094
>WP_010895767.1 MULTISPECIES: ATP-binding protein [Serratia]
MPIFNFRDEDALGKVASVDTTNVIVDVENVEQLKRLQVNHLAVLQSSRPGQHLIGLITQVTRKRGIEDIA
NDGISEQTSELNLCKIALIGTMLDRDGERENVFRRTLESVPEIDANCFSLEGQNLTNFMRTLSSVSADGN
ALTLGKYTLDEHAIAYLNGNKFFQRHAFIGGSTGSGKSWTTAKIIEQMAGLSTANAIVFDLHGEYSPLVG
KGIQHFKVAGPADVEAQRTIDNGVLYLPYWLLSYEALVAMFVDRSDQNAPNQAMIMSREINQAKRKYLED
GDHKEILKHFTVDSPVPFDLDVLMGRLNEINVEMVPGASQGKEKQGDFFGKLARMISRLENKISDRRLGF
IFSGGGDVLNFSWLEKFTTAVLGSSEENGNAGIKIINFSEVPSDVLPLIVSLVARVTFSVQQWTPSELRH
PIALLCDEAHLYMPQRNMAESADDISLDIFERIAKEGRKYGVSLIVISQRPSEVNKTMLSQCSNFVSMRL
TNAEDQGVIKRLLPDSLGGFSDILPTLDTGEALVVGDASLLPSRIRIDEPINKPNSGTVNFWDEWQKPVK
DKRLLIAVENWRKQNIQ
MPIFNFRDEDALGKVASVDTTNVIVDVENVEQLKRLQVNHLAVLQSSRPGQHLIGLITQVTRKRGIEDIA
NDGISEQTSELNLCKIALIGTMLDRDGERENVFRRTLESVPEIDANCFSLEGQNLTNFMRTLSSVSADGN
ALTLGKYTLDEHAIAYLNGNKFFQRHAFIGGSTGSGKSWTTAKIIEQMAGLSTANAIVFDLHGEYSPLVG
KGIQHFKVAGPADVEAQRTIDNGVLYLPYWLLSYEALVAMFVDRSDQNAPNQAMIMSREINQAKRKYLED
GDHKEILKHFTVDSPVPFDLDVLMGRLNEINVEMVPGASQGKEKQGDFFGKLARMISRLENKISDRRLGF
IFSGGGDVLNFSWLEKFTTAVLGSSEENGNAGIKIINFSEVPSDVLPLIVSLVARVTFSVQQWTPSELRH
PIALLCDEAHLYMPQRNMAESADDISLDIFERIAKEGRKYGVSLIVISQRPSEVNKTMLSQCSNFVSMRL
TNAEDQGVIKRLLPDSLGGFSDILPTLDTGEALVVGDASLLPSRIRIDEPINKPNSGTVNFWDEWQKPVK
DKRLLIAVENWRKQNIQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 8862 | GenBank | WP_234591458 |
| Name | trbC_HKJ47_RS00605_pADAP |
UniProt ID | _ |
| Length | 725 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 725 a.a. Molecular weight: 81497.36 Da Isoelectric Point: 8.5425
>WP_234591458.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Serratia]
MSRPVEVDPRRVRRPLGYTFINDALLSPMGVQLSLLAGLIAGFVMPATLLLTVPGLLLLVMLFADRPFRM
PLRMPTDIGGMDLTTEREMPKYRKGLGGFFRYVVRTRKYMPAAGVMCLGYARGKGLARELWLTLDDALRH
MLLLATTGSGKTEALLSVFLNSICWGRGICYSDGKGQNTLAFAMWSLARRFGREDDFYVLNFMTGGTDKL
LQLLLNDKKRQPSNTINLFGTANTTFIIQLMESMLPPAGSGDQGWQDKAKSMLSALVYAIYYKCKREKRR
ISQKVIQEYLPLRKLADLYQEAKRDGWHREAYNPLENYFNTLAGFRIELISKPSEWEQGVYDQHGYLIQQ
FNRMLTMFNDLYGHIFSTDAGDIDIEDVLHNDRILCTTIPALELSKGEASNIGKLYISAIRMTMARDLGC
ELEGMMNDVLIVKKYSGKFPYPIAMDELGAYFGPGMDNLASQMRSLGYMLIVSAQDIQRFIAEHKGEYMT
VNANLLTKWFMTLQDEKDTFELARITGGKGYYAELGSVEQAGGFITPNYEDAANNYIREKDRLDLGDLKD
MSPGEGMISFKSALVPSNAIYIPDDQKMTSALPMRINRFIDVETPTEAELFALNPSLKRKLPPTAQEVDG
ILQRLDLAVDTTASVGIMDPVLKRVAAVALDLDNRADVSYSPAQRGVLLFEAAREALHKNKRKWRQLPTP
PKPIRVSKEVAQSLANAGTNEITFR
MSRPVEVDPRRVRRPLGYTFINDALLSPMGVQLSLLAGLIAGFVMPATLLLTVPGLLLLVMLFADRPFRM
PLRMPTDIGGMDLTTEREMPKYRKGLGGFFRYVVRTRKYMPAAGVMCLGYARGKGLARELWLTLDDALRH
MLLLATTGSGKTEALLSVFLNSICWGRGICYSDGKGQNTLAFAMWSLARRFGREDDFYVLNFMTGGTDKL
LQLLLNDKKRQPSNTINLFGTANTTFIIQLMESMLPPAGSGDQGWQDKAKSMLSALVYAIYYKCKREKRR
ISQKVIQEYLPLRKLADLYQEAKRDGWHREAYNPLENYFNTLAGFRIELISKPSEWEQGVYDQHGYLIQQ
FNRMLTMFNDLYGHIFSTDAGDIDIEDVLHNDRILCTTIPALELSKGEASNIGKLYISAIRMTMARDLGC
ELEGMMNDVLIVKKYSGKFPYPIAMDELGAYFGPGMDNLASQMRSLGYMLIVSAQDIQRFIAEHKGEYMT
VNANLLTKWFMTLQDEKDTFELARITGGKGYYAELGSVEQAGGFITPNYEDAANNYIREKDRLDLGDLKD
MSPGEGMISFKSALVPSNAIYIPDDQKMTSALPMRINRFIDVETPTEAELFALNPSLKRKLPPTAQEVDG
ILQRLDLAVDTTASVGIMDPVLKRVAAVALDLDNRADVSYSPAQRGVLLFEAAREALHKNKRKWRQLPTP
PKPIRVSKEVAQSLANAGTNEITFR
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 691..14099
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HKJ47_RS00010 | 691..1365 | - | 675 | WP_010895742 | DotI/IcmL/TraM family protein | traM |
| HKJ47_RS00015 | 1456..1947 | - | 492 | WP_010895743 | hypothetical protein | traL |
| HKJ47_RS00020 | 1957..5193 | - | 3237 | WP_010895744 | LPD7 domain-containing protein | - |
| HKJ47_RS00025 | 5336..5602 | - | 267 | WP_010895745 | IcmT/TraK family protein | traK |
| HKJ47_RS00030 | 5602..6759 | - | 1158 | WP_010895746 | plasmid transfer ATPase TraJ | virB11 |
| HKJ47_RS00035 | 6949..7737 | - | 789 | WP_010895747 | type IV secretory system conjugative DNA transfer family protein | traI |
| HKJ47_RS00040 | 7734..8192 | - | 459 | WP_010895748 | DotD/TraH family lipoprotein | - |
| HKJ47_RS00045 | 8230..9663 | - | 1434 | WP_010895749 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HKJ47_RS00050 | 9660..10322 | - | 663 | WP_169558203 | prepilin peptidase | - |
| HKJ47_RS00055 | 10328..10813 | - | 486 | WP_010895751 | lytic transglycosylase domain-containing protein | virB1 |
| HKJ47_RS00060 | 10868..11461 | - | 594 | WP_010895752 | type 4 pilus major pilin | - |
| HKJ47_RS00065 | 11497..12609 | - | 1113 | WP_241389818 | type II secretion system F family protein | - |
| HKJ47_RS00070 | 12609..14099 | - | 1491 | WP_010895754 | GspE/PulE family protein | virB11 |
| HKJ47_RS00075 | 14105..14689 | - | 585 | WP_010895755 | type IV pilus biogenesis protein PilP | - |
| HKJ47_RS00080 | 14676..15986 | - | 1311 | WP_010895756 | type 4b pilus protein PilO2 | - |
| HKJ47_RS00085 | 15990..17624 | - | 1635 | WP_010895757 | PilN family type IVB pilus formation outer membrane protein | - |
| HKJ47_RS00090 | 17649..18074 | - | 426 | WP_010895758 | type IV pilus biogenesis protein PilM | - |
| HKJ47_RS00095 | 18074..19087 | - | 1014 | WP_010895759 | toxin co-regulated pilus biosynthesis Q family protein | - |
Region 2: 139650..153518
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HKJ47_RS00595 | 136425..136748 | - | 324 | WP_010895860 | hypothetical protein | - |
| HKJ47_RS00600 | 136758..137294 | - | 537 | WP_234591459 | phospholipase D family protein | - |
| HKJ47_RS00605 | 137435..139612 | - | 2178 | WP_234591458 | F-type conjugative transfer protein TrbC | - |
| HKJ47_RS00610 | 139650..140648 | - | 999 | WP_010895863 | hypothetical protein | trbB |
| HKJ47_RS00615 | 140661..141971 | - | 1311 | WP_010895864 | hypothetical protein | trbA |
| HKJ47_RS00620 | 142109..142690 | - | 582 | WP_010895865 | hypothetical protein | - |
| HKJ47_RS00625 | 142687..144981 | - | 2295 | WP_234591456 | DotA/TraY family protein | traY |
| HKJ47_RS00630 | 144896..145459 | - | 564 | WP_010895867 | conjugal transfer protein TraX | - |
| HKJ47_RS00635 | 145452..146678 | - | 1227 | WP_010895868 | conjugal transfer protein TraW | traW |
| HKJ47_RS00640 | 146819..149893 | - | 3075 | WP_010895869 | TraU | traU |
| HKJ47_RS00645 | 149896..150501 | - | 606 | WP_010895870 | TraT | traT |
| HKJ47_RS00650 | 150587..150988 | - | 402 | WP_010895871 | DUF6750 family protein | traR |
| HKJ47_RS00655 | 151004..151534 | - | 531 | WP_010895872 | conjugal transfer protein TraQ | traQ |
| HKJ47_RS00660 | 151543..152286 | - | 744 | WP_010895873 | hypothetical protein | traP |
| HKJ47_RS00665 | 152286..153518 | - | 1233 | WP_010895874 | conjugal transfer protein TraO | traO |
Host bacterium
| ID | 12453 | GenBank | NC_002523 |
| Plasmid name | pADAP | Incompatibility group | - |
| Plasmid size | 153842 bp | Coordinate of oriT [Strand] | 133975..134057 [+] |
| Host baterium | Serratia entomophila |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |