Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 111044 |
| Name | oriT_pKC-BO-N1-VIM |
| Organism | Kluyvera cryocrescens strain BO64W |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MG228427 (34344..34494 [+], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 12..17, 24..29 (AACCCG..CGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_pKC-BO-N1-VIM
TGGTCGATGTGAACCCGTTTGACCGGGTTATGAATGAGTTGAAAAGTCGTGGCCGCAAGAACGCTCATATCCTGAGCATCCTTCAATTCGACTGGCCCGCATCGGAGGTCATCATCGAGAAGCTGAGCTGTTACATCACAGATGGGATTAA
TGGTCGATGTGAACCCGTTTGACCGGGTTATGAATGAGTTGAAAAGTCGTGGCCGCAAGAACGCTCATATCCTGAGCATCCTTCAATTCGACTGGCCCGCATCGGAGGTCATCATCGAGAAGCTGAGCTGTTACATCACAGATGGGATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file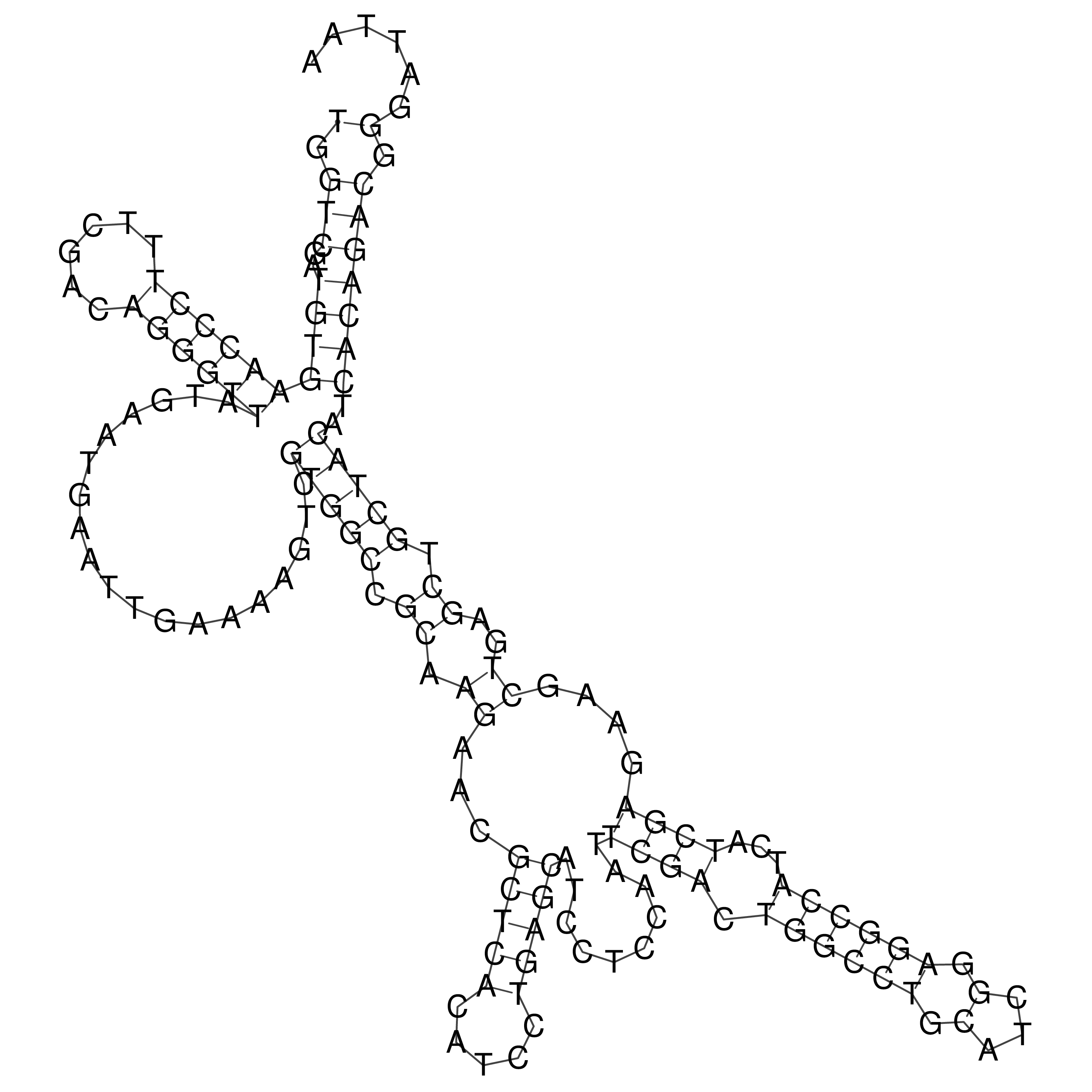
Relaxase
| ID | 7263 | GenBank | WP_058662807 |
| Name | mobH_HTQ50_RS00225_pKC-BO-N1-VIM |
UniProt ID | A0A221KLA3 |
| Length | 987 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 987 a.a. Molecular weight: 109306.91 Da Isoelectric Point: 4.9372
>WP_058662807.1 MULTISPECIES: MobH family relaxase [Gammaproteobacteria]
MLKALHKLFGGRSGAIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWATQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAYRHGIDRYFIRWRDKRHKRHEQFSL
LAWHRIIPAETQEFLSKSGPSIIEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVNEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISQDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGPVKILMLRLESNDLVFTTEPPAAVTGEVVGDVEDAEIE
FVDPEEADEDQAEGDAGLNNDMLAAELEAEKALAGLGFGDAMAMLKSTSDIAEEKPEQEDTESTELPKPD
ASKKGKQQSKPGKARAEKQPQKPEVKEDLSPQDIAKNAPPLANDDPLQALKDVGGGLGDIDFPFDVFNAS
AETACTDATNLAIPDITEPVKQEEQPKPDFVPQEQNSLQNGDFPIFSDSDEPPSWAIEPLPMLTDTPEQT
IPAPDMQHAGKPKELEKDAKTLLSEMLAGYGEASTLLEQAIMPVLEGKTTLGEVLCLMKGQAVILYPEGA
RSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLVDQLSDAVVAAIKDAEASMGGYQEAFELV
SPPGSGVSKNKSAPKQQSRKKTQQQKPEVDAGAGSEQKTKDKSLQQQPKEKQGNVASLVEEQKRKPVQDQ
EKQNVARLPKREAQPVASKPKVEHERELGHVEVRERDEPEVREFEPPKAKTNPKDINEEDFLPSGVTPEK
ALLMLKDMIQKRSGRWLVTPVLEEDGCLVTSVKAFDMIAGENVGISKHILYGLLSRAQRRPLLKKRQGKL
YLEVDKA
MLKALHKLFGGRSGAIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWATQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAYRHGIDRYFIRWRDKRHKRHEQFSL
LAWHRIIPAETQEFLSKSGPSIIEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVNEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISQDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGPVKILMLRLESNDLVFTTEPPAAVTGEVVGDVEDAEIE
FVDPEEADEDQAEGDAGLNNDMLAAELEAEKALAGLGFGDAMAMLKSTSDIAEEKPEQEDTESTELPKPD
ASKKGKQQSKPGKARAEKQPQKPEVKEDLSPQDIAKNAPPLANDDPLQALKDVGGGLGDIDFPFDVFNAS
AETACTDATNLAIPDITEPVKQEEQPKPDFVPQEQNSLQNGDFPIFSDSDEPPSWAIEPLPMLTDTPEQT
IPAPDMQHAGKPKELEKDAKTLLSEMLAGYGEASTLLEQAIMPVLEGKTTLGEVLCLMKGQAVILYPEGA
RSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLVDQLSDAVVAAIKDAEASMGGYQEAFELV
SPPGSGVSKNKSAPKQQSRKKTQQQKPEVDAGAGSEQKTKDKSLQQQPKEKQGNVASLVEEQKRKPVQDQ
EKQNVARLPKREAQPVASKPKVEHERELGHVEVRERDEPEVREFEPPKAKTNPKDINEEDFLPSGVTPEK
ALLMLKDMIQKRSGRWLVTPVLEEDGCLVTSVKAFDMIAGENVGISKHILYGLLSRAQRRPLLKKRQGKL
YLEVDKA
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A221KLA3 |
T4CP
| ID | 8268 | GenBank | WP_000178858 |
| Name | traD_HTQ50_RS00230_pKC-BO-N1-VIM |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69248.76 Da Isoelectric Point: 7.1093
>WP_000178858.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWMAASCAALAVEQVSTMPSEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPEDMWLGSGFLWENRHAQRVFEILKRDWTSIVGKESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVLTHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVKPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNTDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWMAASCAALAVEQVSTMPSEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPEDMWLGSGFLWENRHAQRVFEILKRDWTSIVGKESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVLTHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVKPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNTDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 8269 | GenBank | WP_051479468 |
| Name | traC_HTQ50_RS00310_pKC-BO-N1-VIM |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 92523.78 Da Isoelectric Point: 5.9204
>WP_051479468.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MIITIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDQSVGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTSLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRIFAKTSKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQHLAQIRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYDDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSQKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSTFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSDGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRKQAA
MIITIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDQSVGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTSLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRIFAKTSKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQHLAQIRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYDDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSQKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSTFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSDGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRKQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 32428..57532
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTQ50_RS00215 | 28468..28701 | - | 234 | WP_039272424 | hypothetical protein | - |
| HTQ50_RS00220 | 28683..29165 | - | 483 | WP_228279876 | hypothetical protein | - |
| HTQ50_RS00225 | 29468..32431 | + | 2964 | WP_058662807 | MobH family relaxase | - |
| HTQ50_RS00230 | 32428..34293 | + | 1866 | WP_000178858 | conjugative transfer system coupling protein TraD | virb4 |
| HTQ50_RS00235 | 34304..34888 | + | 585 | WP_000332867 | hypothetical protein | - |
| HTQ50_RS00240 | 34845..35474 | + | 630 | WP_000743448 | DUF4400 domain-containing protein | tfc7 |
| HTQ50_RS00245 | 35500..35931 | + | 432 | WP_228279870 | hypothetical protein | - |
| HTQ50_RS00250 | 35941..36318 | + | 378 | WP_000869260 | hypothetical protein | - |
| HTQ50_RS00255 | 36318..36980 | + | 663 | WP_039272419 | hypothetical protein | - |
| HTQ50_RS00260 | 37159..37302 | + | 144 | WP_168199038 | hypothetical protein | - |
| HTQ50_RS00265 | 37315..37662 | + | 348 | WP_039272417 | hypothetical protein | - |
| HTQ50_RS00270 | 37805..38086 | + | 282 | WP_000805626 | type IV conjugative transfer system protein TraL | traL |
| HTQ50_RS00275 | 38083..38709 | + | 627 | WP_039272415 | TraE/TraK family type IV conjugative transfer system protein | traE |
| HTQ50_RS00280 | 38693..39610 | + | 918 | WP_012766326 | type-F conjugative transfer system secretin TraK | traK |
| HTQ50_RS00285 | 39607..40923 | + | 1317 | WP_039272412 | TraB/VirB10 family protein | traB |
| HTQ50_RS00290 | 40920..41498 | + | 579 | WP_039272410 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTQ50_RS00295 | 41502..41894 | + | 393 | WP_012766327 | TraA family conjugative transfer protein | - |
| HTQ50_RS00300 | 42095..47581 | + | 5487 | WP_039272407 | Ig-like domain-containing protein | - |
| HTQ50_RS00305 | 47730..48437 | + | 708 | WP_001259344 | DsbC family protein | - |
| HTQ50_RS00310 | 48434..50881 | + | 2448 | WP_051479468 | type IV secretion system protein TraC | virb4 |
| HTQ50_RS00315 | 50896..51213 | + | 318 | WP_000364398 | hypothetical protein | - |
| HTQ50_RS00320 | 51210..51740 | + | 531 | WP_000672456 | S26 family signal peptidase | - |
| HTQ50_RS00325 | 51703..52971 | + | 1269 | WP_039272404 | TrbC family F-type conjugative pilus assembly protein | traW |
| HTQ50_RS00330 | 52968..53645 | + | 678 | WP_039272402 | EAL domain-containing protein | - |
| HTQ50_RS00335 | 53651..54643 | + | 993 | WP_236901774 | TraU family protein | traU |
| HTQ50_RS00340 | 54794..57532 | + | 2739 | WP_058662805 | conjugal transfer mating pair stabilization protein TraN | traN |
| HTQ50_RS00345 | 57571..58431 | - | 861 | WP_000679674 | hypothetical protein | - |
| HTQ50_RS00350 | 58543..59193 | - | 651 | WP_000796641 | hypothetical protein | - |
| HTQ50_RS00355 | 59485..59805 | + | 321 | WP_012766334 | hypothetical protein | - |
| HTQ50_RS00360 | 60116..60301 | + | 186 | WP_001186892 | hypothetical protein | - |
| HTQ50_RS00365 | 60522..61490 | + | 969 | WP_001553257 | AAA family ATPase | - |
| HTQ50_RS00370 | 61631..62407 | + | 777 | WP_228279871 | hypothetical protein | - |
Host bacterium
| ID | 11479 | GenBank | NZ_MG228427 |
| Plasmid name | pKC-BO-N1-VIM | Incompatibility group | IncA/C |
| Plasmid size | 170352 bp | Coordinate of oriT [Strand] | 34344..34494 [+] |
| Host baterium | Kluyvera cryocrescens strain BO64W |
Cargo genes
| Drug resistance gene | dfrA14, mph(A), sul1, qacE, catB2, ant(3'')-Ia, aph(3')-XV, blaVIM-1, blaSHV-12, blaACC-1, qnrS1, aph(6)-Id, aph(3'')-Ib, sul2 |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merA, merD |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |