Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 110635 |
| Name | oriT_pSkuCCBAU71714a |
| Organism | Sinorhizobium kummerowiae strain CCBAU 71714 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP120366 (1336541..1336569 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSkuCCBAU71714a
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file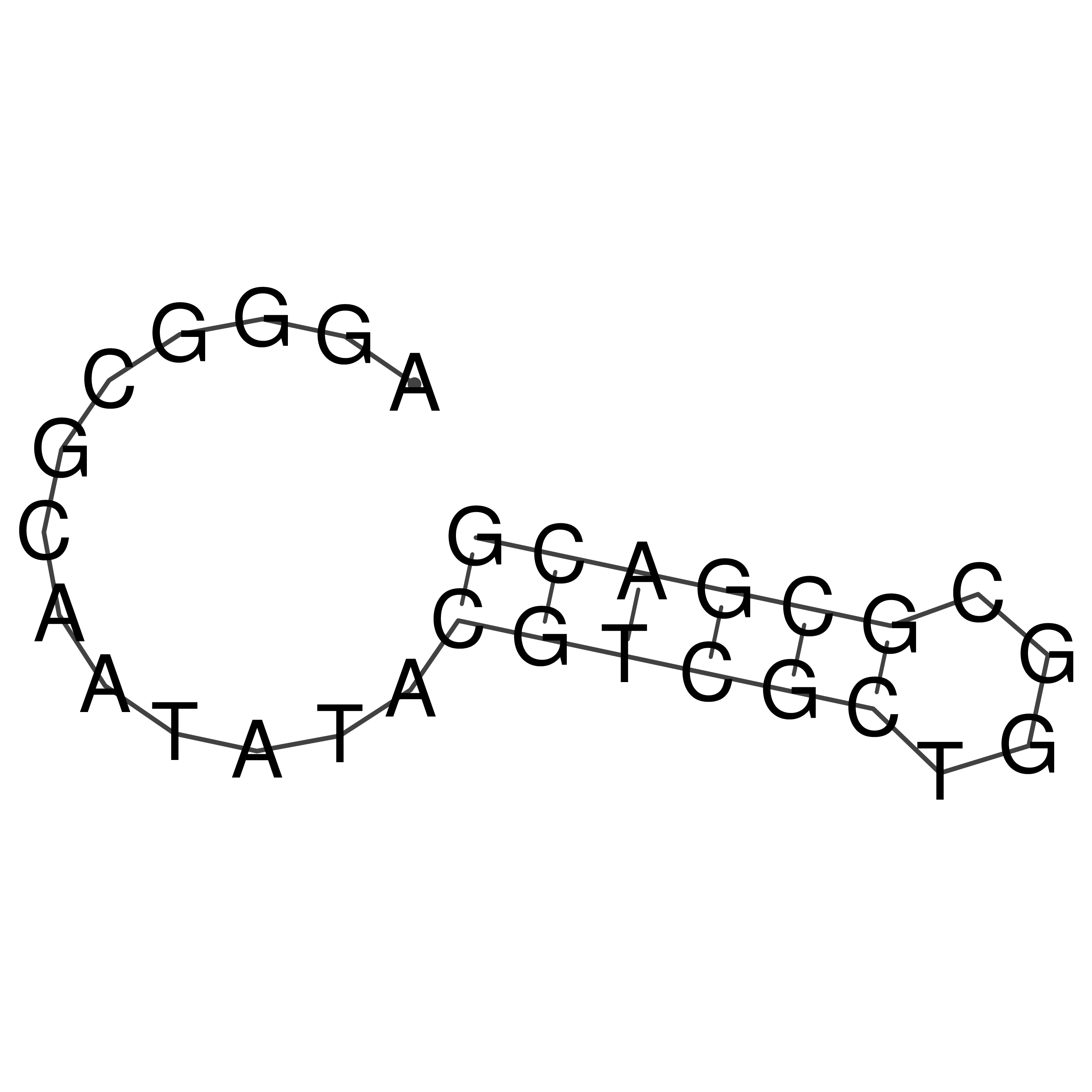
Relaxase
| ID | 7075 | GenBank | WP_284718752 |
| Name | traA_PZL22_RS30995_pSkuCCBAU71714a |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170001.88 Da Isoelectric Point: 9.5526
>WP_284718752.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium kummerowiae]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRHEMADRLLAEPGLLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDELVLLKPQQIDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSDARAAAAVRSIETADTEKPFRLDLEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRDLLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFIEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGREVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPKWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGAAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPERLGRLRGSELVVDGRAARDERTAATA
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDIAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRIQSSEKIVSERQRRVIDRARGVTR
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRHEMADRLLAEPGLLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDELVLLKPQQIDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSDARAAAAVRSIETADTEKPFRLDLEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRDLLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFIEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGREVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPKWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGAAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPERLGRLRGSELVVDGRAARDERTAATA
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDIAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRIQSSEKIVSERQRRVIDRARGVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 8023 | GenBank | WP_127701843 |
| Name | virD4_PZL22_RS26935_pSkuCCBAU71714a |
UniProt ID | _ |
| Length | 563 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 563 a.a. Molecular weight: 62957.23 Da Isoelectric Point: 9.1751
>WP_127701843.1 MULTISPECIES: type IV secretion system ATPase VirD4 [Sinorhizobium]
MISSKTRPLSIAGSIMCSLAVGFCGASAYSTFRFGFDGRALMTFDILAFWYETPFYLGYTTLFFYRGLAV
VVLTSGAILFVQQMVSVRDRQHHGTARWARVDEMRRAGYLQRYSRISGPVFGKTSGPFWSDYYLTNSEQP
HSLIVAPTRAGKGVGIVIPTLLTYEGSVIALDVKGELFDLTSRARKARGDSVFKLAPLDPERRTNCYNPL
LDIVALPSERQFTEARRLAANLIATKGQSAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGEKY
SLFARLAQETQNKEAQRIFDEMASNDTKILTSYTSVLGDGGLNLWADPLIKAATSRSDFSIYDLRRRRTC
IYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMFIIQSLSALTGTYEESGKQNFLSNTGVQVFMATADDETPVYISKAIGEYTFQARSTSYTQSLTFDRNI
QNSDQGAPLLRPEQVRLLPDKYQIVLIKGQPPLQLRKVRYYSDRALKRIFDSQTGRLPEPAPLMIADERF
SHV
MISSKTRPLSIAGSIMCSLAVGFCGASAYSTFRFGFDGRALMTFDILAFWYETPFYLGYTTLFFYRGLAV
VVLTSGAILFVQQMVSVRDRQHHGTARWARVDEMRRAGYLQRYSRISGPVFGKTSGPFWSDYYLTNSEQP
HSLIVAPTRAGKGVGIVIPTLLTYEGSVIALDVKGELFDLTSRARKARGDSVFKLAPLDPERRTNCYNPL
LDIVALPSERQFTEARRLAANLIATKGQSAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGEKY
SLFARLAQETQNKEAQRIFDEMASNDTKILTSYTSVLGDGGLNLWADPLIKAATSRSDFSIYDLRRRRTC
IYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMFIIQSLSALTGTYEESGKQNFLSNTGVQVFMATADDETPVYISKAIGEYTFQARSTSYTQSLTFDRNI
QNSDQGAPLLRPEQVRLLPDKYQIVLIKGQPPLQLRKVRYYSDRALKRIFDSQTGRLPEPAPLMIADERF
SHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 8024 | GenBank | WP_122102935 |
| Name | traG_PZL22_RS30980_pSkuCCBAU71714a |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70239.23 Da Isoelectric Point: 9.3854
>WP_122102935.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Sinorhizobium]
MALKAKPHPSLLVILFPVAVTTAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVVGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERCRVDKDIVHELPFDPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEEIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
MALKAKPHPSLLVILFPVAVTTAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVVGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERCRVDKDIVHELPFDPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEEIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 21451..31026
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PZL22_RS24460 (PZL22_004896) | 17768..18055 | - | 288 | WP_013845449 | chaperone NapD | - |
| PZL22_RS24465 (PZL22_004897) | 18048..18548 | - | 501 | WP_088201148 | ferredoxin-type protein NapF | - |
| PZL22_RS24470 (PZL22_004898) | 18548..18733 | - | 186 | WP_088198221 | periplasmic nitrate reductase, NapE protein | - |
| PZL22_RS24475 (PZL22_004899) | 19264..19875 | + | 612 | WP_127525415 | TetR/AcrR family transcriptional regulator | - |
| PZL22_RS24480 (PZL22_004900) | 19914..21140 | + | 1227 | WP_127634296 | MFS transporter | - |
| PZL22_RS24485 (PZL22_004901) | 21451..22440 | - | 990 | WP_127525413 | P-type DNA transfer ATPase VirB11 | virB11 |
| PZL22_RS24490 (PZL22_004902) | 22451..23623 | - | 1173 | WP_013845455 | type IV secretion system protein VirB10 | virB10 |
| PZL22_RS24495 (PZL22_004903) | 23633..24487 | - | 855 | WP_014531548 | P-type conjugative transfer protein VirB9 | virB9 |
| PZL22_RS24500 (PZL22_004904) | 24487..25158 | - | 672 | WP_013845457 | virB8 family protein | virB8 |
| PZL22_RS24505 (PZL22_004905) | 25163..25477 | - | 315 | WP_013845458 | hypothetical protein | - |
| PZL22_RS24510 (PZL22_004906) | 25474..26409 | - | 936 | WP_127525411 | type IV secretion system protein | virB6 |
| PZL22_RS24515 (PZL22_004907) | 26432..26647 | - | 216 | WP_014531545 | EexN family lipoprotein | - |
| PZL22_RS24520 (PZL22_004908) | 26644..27342 | - | 699 | WP_127634297 | P-type DNA transfer protein VirB5 | virB5 |
| PZL22_RS24525 (PZL22_004909) | 27342..29738 | - | 2397 | WP_192925931 | VirB4 family type IV secretion system protein | virb4 |
| PZL22_RS24530 (PZL22_004910) | 29713..30054 | - | 342 | WP_017264162 | type IV secretion system protein VirB3 | virB3 |
| PZL22_RS24535 (PZL22_004911) | 30059..30358 | - | 300 | WP_088196212 | TrbC/VirB2 family protein | virB2 |
| PZL22_RS24540 (PZL22_004912) | 30355..31026 | - | 672 | WP_127525410 | lytic transglycosylase domain-containing protein | virB1 |
| PZL22_RS24545 (PZL22_004913) | 31030..31548 | - | 519 | WP_088196214 | hypothetical protein | - |
| PZL22_RS24550 (PZL22_004914) | 31645..32013 | + | 369 | WP_017264159 | transcriptional regulator | - |
| PZL22_RS24555 (PZL22_004915) | 32128..32511 | - | 384 | WP_234837683 | hypothetical protein | - |
| PZL22_RS24560 (PZL22_004916) | 32586..33233 | - | 648 | WP_088196216 | dihydrofolate reductase family protein | - |
| PZL22_RS24565 (PZL22_004917) | 33283..34131 | - | 849 | WP_127525409 | alpha/beta fold hydrolase | - |
Host bacterium
| ID | 11070 | GenBank | NZ_CP120366 |
| Plasmid name | pSkuCCBAU71714a | Incompatibility group | - |
| Plasmid size | 1418903 bp | Coordinate of oriT [Strand] | 1336541..1336569 [+] |
| Host baterium | Sinorhizobium kummerowiae strain CCBAU 71714 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | ncrA |
| Degradation gene | - |
| Symbiosis gene | fixB, fixA, fixN, fixO, fixG, fixS, nodD, fixU, nifB, fixX, fixC, nifH, nifD, nifK, nifE, nifX, nodH, nodF, nodE, nodJ, nodI, nodC, nodB, nodA, nifN, nodM |
| Anti-CRISPR | AcrIIA9 |