Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 110119 |
| Name | oriT_pHN21SC92-3 |
| Organism | Citrobacter portucalensis strain GD21SC92T |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP089440 (5694..6309 [-], 616 nt) |
| oriT length | 616 nt |
| IRs (inverted repeats) | 599..604, 609..614 (ATGCCC..GGGCAT) 526..532, 538..544 (CCTCCCG..CGGGAGG) 462..467, 471..476 (GTTCGC..GCGAAC) 52..58, 62..68 (CATTATC..GATAATG) 40..45, 54..59 (TGATAA..TTATCA) |
| Location of nic site | 322..323 |
| Conserved sequence flanking the nic site |
TCCTGCATCG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 616 nt
>oriT_pHN21SC92-3
TCTTACTTCTTTGCGTAGCTGTTAAATACAGCGTTGTTTTGATAAAATCATCATTATCATCGATAATGCTTTCTTCAATTTTTTTATCCTTACTCTTTAATAAAGCACTTGCTAATAACTTCATACCTTTTGCAACTGTCAAATTTGGTTCATCAGGGTAAATGCTTTTAAGGCATACTAACAAATAATCATGGTCTTCATCTTCAACTCTAAACTGAATTTTTTTCATCATAACTCCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGCTGAAAAATAACGTCGAATGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAATCTGGCGCCGCTCATAAATTGTGCTTGAGCATCAACAAAAATGCAAAAAGGCTGATGTTGTCATGGCTGAACATACGTACGTCTGCGACCGGAAACCCGATCAATGCGGTAAGGACAGTGCCGTCAGGTTCGCTCCGCGAACGGTCTGGCAGGAAGCGCAAGCGCAGTCCCGCGCAATATATCAGAAAGCACCTCCCGAAATACGGGAGGGCTTCGGCTATTCAGCTAAGGTTGCTCTGTTCAGGTTGCTGAGTGTAGCCTTCCATGCCCTGACGGGCATCA
TCTTACTTCTTTGCGTAGCTGTTAAATACAGCGTTGTTTTGATAAAATCATCATTATCATCGATAATGCTTTCTTCAATTTTTTTATCCTTACTCTTTAATAAAGCACTTGCTAATAACTTCATACCTTTTGCAACTGTCAAATTTGGTTCATCAGGGTAAATGCTTTTAAGGCATACTAACAAATAATCATGGTCTTCATCTTCAACTCTAAACTGAATTTTTTTCATCATAACTCCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGCTGAAAAATAACGTCGAATGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAATCTGGCGCCGCTCATAAATTGTGCTTGAGCATCAACAAAAATGCAAAAAGGCTGATGTTGTCATGGCTGAACATACGTACGTCTGCGACCGGAAACCCGATCAATGCGGTAAGGACAGTGCCGTCAGGTTCGCTCCGCGAACGGTCTGGCAGGAAGCGCAAGCGCAGTCCCGCGCAATATATCAGAAAGCACCTCCCGAAATACGGGAGGGCTTCGGCTATTCAGCTAAGGTTGCTCTGTTCAGGTTGCTGAGTGTAGCCTTCCATGCCCTGACGGGCATCA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file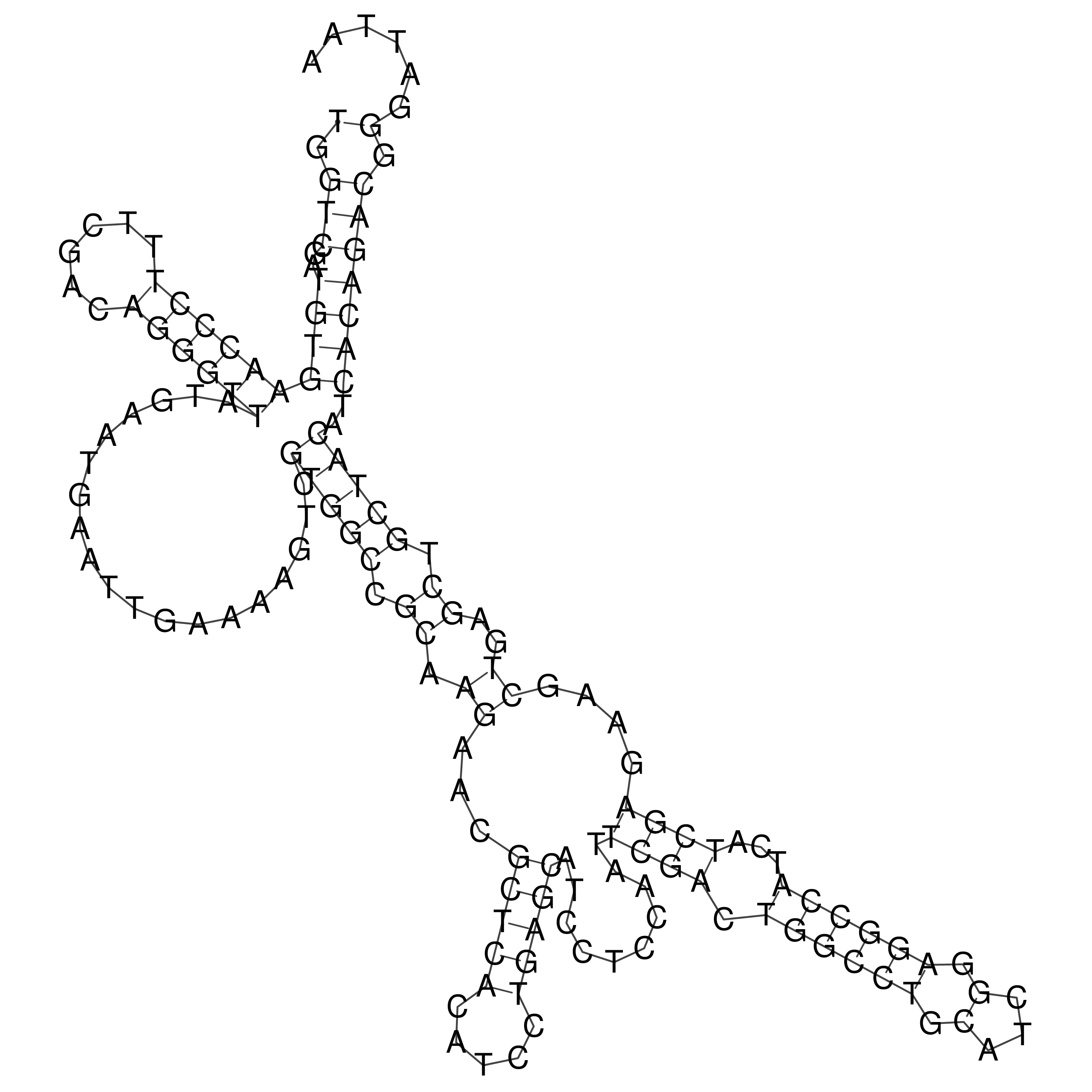
T4CP
| ID | 7395 | GenBank | WP_178970148 |
| Name | t4cp2_LT980_RS27040_pHN21SC92-3 |
UniProt ID | _ |
| Length | 919 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 919 a.a. Molecular weight: 104740.75 Da Isoelectric Point: 6.1962
>WP_178970148.1 MULTISPECIES: VirB3 family type IV secretion system protein [Enterobacteriaceae]
MSTVFKGLTRPALIRGLGVPLYPFLAMCVLVVLLGVWIDDLMYFLILPGWFAIKRVTKIDERFFDLLYLR
AVVKGHPLANKRFSAVHYAGSQYDEVDISKVDNFMKLKDQASIEALIPYSSHITDDLVVTKNRDLLATWQ
VDGAYFECVDDEDLTLLTDQLNTLIRSFEGRAITFYTHRVRVRKEVKVGFNSKIPFVNRVMNDYYSSLSE
PEYFENRLYLTICYKPFNIEDKVSHFISKNKKENNIFDEPINDMNEICGRLDTYLSRFHAHRLGLVEENG
RVFSDQLSLFQYLLSGKWQKVRVTNSPFYTYLGGKDLFFGNDAGQITASQNARYFRSIEIKDYFQETDAG
IFDALMYLPVEYVFTSSFTSMDKQAAVKALDDQIDKLELTDDAAKSLLADLRVGLDMVSSGYNSFGKCHQ
TLIVFADTPERLVKDTNIVTTTLEDLGLIVTYSTLSLGAAFFSQLPGNYNLRPRLSMLSSLNFAEMESFH
NFFHGKATGNTWGNSLMALRGSGNDVYHLNYHMTTENINFFGKNPTLGHCEILGTSNVGKTVLMMIMSYA
AQQFGTPESFPENRKVRKLTTVFFDKDRAGEVGIRAIGGAYFRVKGGEPTGWNPAALPPTKRNIAFVKDI
IRLICKLNGNTVDDYQHSLISSAVDRLMQREDRSFPISKLIPLIMEPDDTETKRHGLKARLRAWKQGGEY
GWLLDNASDSFDVAHLDVFGIDGTEFLDDKVVAPVASFYLIYRVTMLADGRRLLIYMDEFWQWINNDAFK
DFVYNKLKTGRKLDMVLVPATQSPDELIKSPIAAAVREQCATHIYLANPKAKRNEYVDELQVRDLYFDKI
KAIDPLSRQFLVVKNPQRQGERDDFAAFAKLDLGKAAYYLPILSASAEQLELFDEIWSEGMAPEEWIDTY
LERANRGVK
MSTVFKGLTRPALIRGLGVPLYPFLAMCVLVVLLGVWIDDLMYFLILPGWFAIKRVTKIDERFFDLLYLR
AVVKGHPLANKRFSAVHYAGSQYDEVDISKVDNFMKLKDQASIEALIPYSSHITDDLVVTKNRDLLATWQ
VDGAYFECVDDEDLTLLTDQLNTLIRSFEGRAITFYTHRVRVRKEVKVGFNSKIPFVNRVMNDYYSSLSE
PEYFENRLYLTICYKPFNIEDKVSHFISKNKKENNIFDEPINDMNEICGRLDTYLSRFHAHRLGLVEENG
RVFSDQLSLFQYLLSGKWQKVRVTNSPFYTYLGGKDLFFGNDAGQITASQNARYFRSIEIKDYFQETDAG
IFDALMYLPVEYVFTSSFTSMDKQAAVKALDDQIDKLELTDDAAKSLLADLRVGLDMVSSGYNSFGKCHQ
TLIVFADTPERLVKDTNIVTTTLEDLGLIVTYSTLSLGAAFFSQLPGNYNLRPRLSMLSSLNFAEMESFH
NFFHGKATGNTWGNSLMALRGSGNDVYHLNYHMTTENINFFGKNPTLGHCEILGTSNVGKTVLMMIMSYA
AQQFGTPESFPENRKVRKLTTVFFDKDRAGEVGIRAIGGAYFRVKGGEPTGWNPAALPPTKRNIAFVKDI
IRLICKLNGNTVDDYQHSLISSAVDRLMQREDRSFPISKLIPLIMEPDDTETKRHGLKARLRAWKQGGEY
GWLLDNASDSFDVAHLDVFGIDGTEFLDDKVVAPVASFYLIYRVTMLADGRRLLIYMDEFWQWINNDAFK
DFVYNKLKTGRKLDMVLVPATQSPDELIKSPIAAAVREQCATHIYLANPKAKRNEYVDELQVRDLYFDKI
KAIDPLSRQFLVVKNPQRQGERDDFAAFAKLDLGKAAYYLPILSASAEQLELFDEIWSEGMAPEEWIDTY
LERANRGVK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 7396 | GenBank | WP_000053826 |
| Name | t4cp2_LT980_RS27080_pHN21SC92-3 |
UniProt ID | _ |
| Length | 611 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 611 a.a. Molecular weight: 69231.07 Da Isoelectric Point: 8.0208
>WP_000053826.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MSLKLPDKGQWAFIILVMCLIAYYTGSVAIYFFNHKTPLYIWKHYDSLLLWRVIIDSSIKSEVRFTALPS
LLSGALASLAAPAFIIWKLNQKDAPLFGDAKFASDSDLKKSKLLKWEKENDTDILVGRYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNMLVKKHSIIALDPKQELWKITSKVREQILGNKVYLLDPFNTKTHKCNPLF
YVDLKSESGAKDLLKLVEILFPSFGLTGAEAHFNNLAGQYWTGLAKLLHFFINFAPEWIEELRVKPVFSI
GTVVDLYSNIDREQVLSNRETFEELAQGNETALFHLKDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQMRREDITVYVGVNAEDMILAYDFLNLFFNLVVEVTLRENPDFDPTLKHDCLLFL
DEFPSIGYMPIIKKGSGYIAGYKLKLLTIYQNISQLNEIYGTEGAKTLLSAHPCRVIYAVSEEDDAKKIS
EKLGYITARSKGSSKTSGKATSRSNSENEAQRALVLPQELGTLDFSEEMIILKGEHPVKAEKALYYLDNY
FMERLMLVSPKLTALTASINRSNKIFGVKGIKYPSKEKMLSVGELESEVLL
MSLKLPDKGQWAFIILVMCLIAYYTGSVAIYFFNHKTPLYIWKHYDSLLLWRVIIDSSIKSEVRFTALPS
LLSGALASLAAPAFIIWKLNQKDAPLFGDAKFASDSDLKKSKLLKWEKENDTDILVGRYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNMLVKKHSIIALDPKQELWKITSKVREQILGNKVYLLDPFNTKTHKCNPLF
YVDLKSESGAKDLLKLVEILFPSFGLTGAEAHFNNLAGQYWTGLAKLLHFFINFAPEWIEELRVKPVFSI
GTVVDLYSNIDREQVLSNRETFEELAQGNETALFHLKDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQMRREDITVYVGVNAEDMILAYDFLNLFFNLVVEVTLRENPDFDPTLKHDCLLFL
DEFPSIGYMPIIKKGSGYIAGYKLKLLTIYQNISQLNEIYGTEGAKTLLSAHPCRVIYAVSEEDDAKKIS
EKLGYITARSKGSSKTSGKATSRSNSENEAQRALVLPQELGTLDFSEEMIILKGEHPVKAEKALYYLDNY
FMERLMLVSPKLTALTASINRSNKIFGVKGIKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 8863..18753
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LT980_RS26990 (LT980_26980) | 4161..4406 | + | 246 | WP_000356546 | hypothetical protein | - |
| LT980_RS26995 (LT980_26985) | 4396..4611 | + | 216 | WP_001180116 | hypothetical protein | - |
| LT980_RS27000 (LT980_26990) | 4704..5033 | + | 330 | WP_000866648 | hypothetical protein | - |
| LT980_RS27005 (LT980_26995) | 5320..5469 | + | 150 | WP_000003880 | hypothetical protein | - |
| LT980_RS27010 (LT980_27000) | 5482..5754 | + | 273 | WP_000160399 | hypothetical protein | - |
| LT980_RS27015 (LT980_27005) | 6081..6626 | + | 546 | WP_038976855 | DNA distortion polypeptide 1 | - |
| LT980_RS27020 (LT980_27010) | 6629..7786 | + | 1158 | WP_000538023 | DNA distortion polypeptide 2 | - |
| LT980_RS27025 (LT980_27015) | 8147..8638 | + | 492 | WP_000872475 | transcription termination/antitermination NusG family protein | - |
| LT980_RS27030 (LT980_27020) | 8863..9507 | + | 645 | WP_000953539 | lytic transglycosylase domain-containing protein | virB1 |
| LT980_RS27035 (LT980_27025) | 9491..9781 | + | 291 | WP_000921916 | TrbC/VirB2 family protein | virB2 |
| LT980_RS27040 (LT980_27030) | 9805..12564 | + | 2760 | WP_178970148 | VirB3 family type IV secretion system protein | virb4 |
| LT980_RS27045 (LT980_27035) | 12566..13303 | + | 738 | WP_000737859 | type IV secretion system protein | - |
| LT980_RS27050 (LT980_27040) | 13313..13573 | + | 261 | WP_001228869 | EexN family lipoprotein | - |
| LT980_RS27055 (LT980_27045) | 13585..14640 | + | 1056 | WP_000235774 | type IV secretion system protein | virB6 |
| LT980_RS27060 (LT980_27050) | 14830..15552 | + | 723 | WP_000394570 | type IV secretion system protein | virB8 |
| LT980_RS27065 (LT980_27055) | 15558..16487 | + | 930 | WP_000776689 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| LT980_RS27070 (LT980_27060) | 16484..17698 | + | 1215 | WP_001295061 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| LT980_RS27075 (LT980_27065) | 17716..18753 | + | 1038 | WP_000217791 | P-type DNA transfer ATPase VirB11 | virB11 |
| LT980_RS27080 (LT980_27070) | 18758..20593 | + | 1836 | WP_000053826 | type IV secretory system conjugative DNA transfer family protein | - |
| LT980_RS27085 (LT980_27075) | 20590..20985 | + | 396 | WP_000733627 | cag pathogenicity island Cag12 family protein | - |
| LT980_RS27090 (LT980_27080) | 20988..21320 | + | 333 | WP_000699980 | hypothetical protein | - |
| LT980_RS27095 (LT980_27085) | 21563..22378 | + | 816 | WP_000018321 | aminoglycoside O-phosphotransferase APH(3')-Ia | - |
| LT980_RS27100 (LT980_27090) | 22532..23440 | - | 909 | WP_000174819 | C45 family peptidase | - |
| LT980_RS27105 (LT980_27095) | 23587..23673 | + | 87 | Protein_29 | micrococcal nuclease | - |
Host bacterium
| ID | 10554 | GenBank | NZ_CP089440 |
| Plasmid name | pHN21SC92-3 | Incompatibility group | IncX1 |
| Plasmid size | 48371 bp | Coordinate of oriT [Strand] | 5694..6309 [-] |
| Host baterium | Citrobacter portucalensis strain GD21SC92T |
Cargo genes
| Drug resistance gene | aph(3')-Ia, qnrS1, dfrA5, tet(A), blaTEM-1B |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |