Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 110112 |
| Name | oriT_pHN21SC1505-2 |
| Organism | Enterobacter roggenkampii strain GD21SC1505 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP091083 (26681..27321 [+], 641 nt) |
| oriT length | 641 nt |
| IRs (inverted repeats) | 597..602, 607..612 (ATGCCC..GGGCAT) 526..532, 537..543 (CCTCCCG..CGGGAGG) 460..465, 469..474 (GTTCGC..GCGAAC) 345..350, 358..363 (ACTGAA..TTCAGT) 52..58, 62..68 (CATTATC..GATAATG) 40..45, 54..59 (TGATAA..TTATCA) |
| Location of nic site | 322..323 |
| Conserved sequence flanking the nic site |
TCCTGCATCG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 641 nt
>oriT_pHN21SC1505-2
TCTTACTTCTTTGCGTATCTGTTAAATACAGCGTTGTTTTGATAAAATCATCATTATCATCGATAATGCTTTCTTCAATTTTTTTATCCTTACTCTTTAATAAAGCACTTGCTAATAACTTCATACCTTTTGCAACTGTCAAATTTGGTTCATCAGGGTAAATGCTTTTAAGGCATACTAACAAATAATCATGGTCTTCATCTTCAACTCTAAACTGAATTTTTTTCATCATAACTCCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGCTGAAAAATAACGTCGAATGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTAGAAAATGCCACAACTGAAATTGTGCTTCAGTATGTACAGAAATGCAAAATCTGAGGGATTTCGTAGCTGAAAGATCGCCAGTCTTCGACCGTAAGGATAGGAGTTGCTGTAAGACCTGTGCGGGGCTGTTCGCTTCGCGAACGGGTCTGGCAGGGGGCGCAAGCGCTGTGCTGTGATATATGCAAAAGAAGCACCTCCCGCAAACGGGAGGGCTTCGGCGAATCGACTATAGTGATCTATTTACCCGGCTGATTGTCGCCTTCTATGCCCTCGCGGGCATCATGCAACCAGCGCCTGAATTTAGTTATA
TCTTACTTCTTTGCGTATCTGTTAAATACAGCGTTGTTTTGATAAAATCATCATTATCATCGATAATGCTTTCTTCAATTTTTTTATCCTTACTCTTTAATAAAGCACTTGCTAATAACTTCATACCTTTTGCAACTGTCAAATTTGGTTCATCAGGGTAAATGCTTTTAAGGCATACTAACAAATAATCATGGTCTTCATCTTCAACTCTAAACTGAATTTTTTTCATCATAACTCCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGCTGAAAAATAACGTCGAATGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTAGAAAATGCCACAACTGAAATTGTGCTTCAGTATGTACAGAAATGCAAAATCTGAGGGATTTCGTAGCTGAAAGATCGCCAGTCTTCGACCGTAAGGATAGGAGTTGCTGTAAGACCTGTGCGGGGCTGTTCGCTTCGCGAACGGGTCTGGCAGGGGGCGCAAGCGCTGTGCTGTGATATATGCAAAAGAAGCACCTCCCGCAAACGGGAGGGCTTCGGCGAATCGACTATAGTGATCTATTTACCCGGCTGATTGTCGCCTTCTATGCCCTCGCGGGCATCATGCAACCAGCGCCTGAATTTAGTTATA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file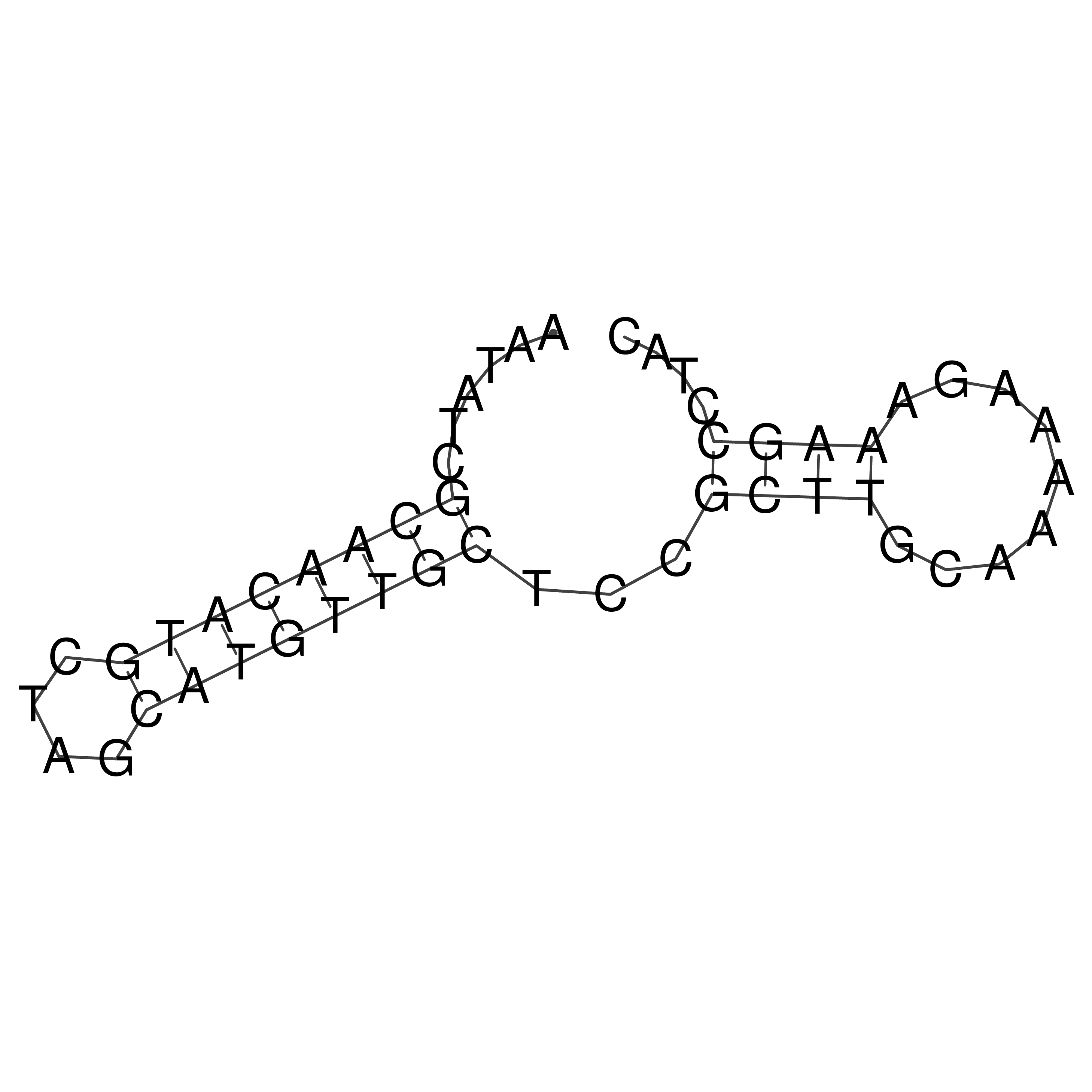
T4CP
| ID | 7388 | GenBank | WP_250670776 |
| Name | t4cp2_L1S38_RS24060_pHN21SC1505-2 |
UniProt ID | _ |
| Length | 611 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 611 a.a. Molecular weight: 69263.11 Da Isoelectric Point: 8.0208
>WP_250670776.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MSLKLPDKGQWAFIILVMCLIVYYTGSVAIYFFNHKTTLYIWKHYDSLLLWRVIIDSSIKSEVRFTALPS
LLSGALASLAAPAFIIWKLNQKDAPLFGDAKFASDSDLKKSKLLKWEKENDTDILVGRYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNMLVKKHSIIALDPKQELWKITSKVREQILGNKVYLLDPFNTKTHKCNPLF
YVDLKSESGAKDLLKLVEILFPSFGLTGAEAHFNNLAGQYWTGLAKLLHFFINFAPEWIEELRVKPVFSI
GTVVDLYSNIDREQVLSNRETFEELAQGNETALFHLKDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQMRREDITVYVGVNAEDMILAYDFLNLFFNLVVEVTLRENPDFDPTLKHDCLLFL
DEFPSIGYMPIIKKGSGYIAGYKLKLLTIYQNISQLNEIYGTEGAKTLLSAHPCRVIYAVSEEDDAKKIS
EKLGYITARSKGSSKTSGKATSRSNSENEAQRALVLPQELGTLDFSEEMIILKGEHPVKAEKALYYLDNY
FMERLMLVSPKLTALTASINRSNKIFGVKGIKYPSKEKMLSVGELESEVLL
MSLKLPDKGQWAFIILVMCLIVYYTGSVAIYFFNHKTTLYIWKHYDSLLLWRVIIDSSIKSEVRFTALPS
LLSGALASLAAPAFIIWKLNQKDAPLFGDAKFASDSDLKKSKLLKWEKENDTDILVGRYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNMLVKKHSIIALDPKQELWKITSKVREQILGNKVYLLDPFNTKTHKCNPLF
YVDLKSESGAKDLLKLVEILFPSFGLTGAEAHFNNLAGQYWTGLAKLLHFFINFAPEWIEELRVKPVFSI
GTVVDLYSNIDREQVLSNRETFEELAQGNETALFHLKDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQMRREDITVYVGVNAEDMILAYDFLNLFFNLVVEVTLRENPDFDPTLKHDCLLFL
DEFPSIGYMPIIKKGSGYIAGYKLKLLTIYQNISQLNEIYGTEGAKTLLSAHPCRVIYAVSEEDDAKKIS
EKLGYITARSKGSSKTSGKATSRSNSENEAQRALVLPQELGTLDFSEEMIILKGEHPVKAEKALYYLDNY
FMERLMLVSPKLTALTASINRSNKIFGVKGIKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 7389 | GenBank | WP_000108725 |
| Name | t4cp2_L1S38_RS24100_pHN21SC1505-2 |
UniProt ID | _ |
| Length | 919 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 919 a.a. Molecular weight: 104758.78 Da Isoelectric Point: 6.1962
>WP_000108725.1 MULTISPECIES: VirB3 family type IV secretion system protein [Enterobacteriaceae]
MSTVFKGLTRPALIRGLGVPLYPFLAMCVLVVLLGVWIDDLMYFLILPGWFAIKRVTKIDERFFDLLYLR
AVVKGHPLANKRFSAVHYAGSQYDEVDISKVDNFMKLKDQASIEALIPYSSHITDDLVVTKNRDLLATWQ
VDGAYFECVDDEDLTLLTDQLNTLIRSFEGRAITFYTHRVRVRKEVKVGFNSKIPFVNRVMNDYYSSLSE
PEYFENRLYLTICYKPFNIEDKVSHFISKNKKENNIFDEPINDMNEICGRLDTYLSRFHAHRLGLVEENG
RVFSDQLSLFQYLLSGKWQKVRVTNSPFYTYLGGKDLFFGNDAGQITASQNARYFRSIEIKDYFQETDAG
IFDALMYLPVEYVFTSSFTSMDKQAAVKALDDQIDKLELTDDAAKSLLADLRVGLDMVSSGYNSFGKCHQ
TLIVFADTPERLVKDTNIVTTTLEDLGLIVTYSTLSLGAAFFSQLPGNYNLRPRLSMLSSLNFAEMESFH
NFFHGKATGNTWGNSLMALRGSGNDVYHLNYHMTTENINFFGKNPTLGHCEILGTSNVGKTVLMMIMSYA
AQQFGTPESFPENRKVRKLTTVFFDKDRAGEVGIRAMGGAYFRVKGGEPTGWNPAALPPTKRNIAFVKDI
IRLICKLNGNTVDDYQHSLISSAVDRLMQREDRSFPISKLIPLIMEPDDTETKRHGLKARLRAWKQGGEY
GWLLDNASDSFDVAHLDVFGIDGTEFLDDKVVAPVASFYLIYRVTMLADGRRLLIYMDEFWQWINNDAFK
DFVYNKLKTGRKLDMVLVPATQSPDELIKSPIAAAVREQCATHIYLANPKAKRNEYVDELQVRDLYFDKI
KAIDPLSRQFLVVKNPQRQGERDDFAAFAKLDLGKAAYYLPILSASAEQLELFDEIWSEGMAPEEWIDTY
LERANRGVK
MSTVFKGLTRPALIRGLGVPLYPFLAMCVLVVLLGVWIDDLMYFLILPGWFAIKRVTKIDERFFDLLYLR
AVVKGHPLANKRFSAVHYAGSQYDEVDISKVDNFMKLKDQASIEALIPYSSHITDDLVVTKNRDLLATWQ
VDGAYFECVDDEDLTLLTDQLNTLIRSFEGRAITFYTHRVRVRKEVKVGFNSKIPFVNRVMNDYYSSLSE
PEYFENRLYLTICYKPFNIEDKVSHFISKNKKENNIFDEPINDMNEICGRLDTYLSRFHAHRLGLVEENG
RVFSDQLSLFQYLLSGKWQKVRVTNSPFYTYLGGKDLFFGNDAGQITASQNARYFRSIEIKDYFQETDAG
IFDALMYLPVEYVFTSSFTSMDKQAAVKALDDQIDKLELTDDAAKSLLADLRVGLDMVSSGYNSFGKCHQ
TLIVFADTPERLVKDTNIVTTTLEDLGLIVTYSTLSLGAAFFSQLPGNYNLRPRLSMLSSLNFAEMESFH
NFFHGKATGNTWGNSLMALRGSGNDVYHLNYHMTTENINFFGKNPTLGHCEILGTSNVGKTVLMMIMSYA
AQQFGTPESFPENRKVRKLTTVFFDKDRAGEVGIRAMGGAYFRVKGGEPTGWNPAALPPTKRNIAFVKDI
IRLICKLNGNTVDDYQHSLISSAVDRLMQREDRSFPISKLIPLIMEPDDTETKRHGLKARLRAWKQGGEY
GWLLDNASDSFDVAHLDVFGIDGTEFLDDKVVAPVASFYLIYRVTMLADGRRLLIYMDEFWQWINNDAFK
DFVYNKLKTGRKLDMVLVPATQSPDELIKSPIAAAVREQCATHIYLANPKAKRNEYVDELQVRDLYFDKI
KAIDPLSRQFLVVKNPQRQGERDDFAAFAKLDLGKAAYYLPILSASAEQLELFDEIWSEGMAPEEWIDTY
LERANRGVK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 14237..24127
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| L1S38_RS24040 (L1S38_24040) | 10457..10702 | - | 246 | WP_001243650 | hypothetical protein | - |
| L1S38_RS24045 (L1S38_24045) | 11117..11587 | - | 471 | WP_025755767 | thermonuclease family protein | - |
| L1S38_RS24050 (L1S38_24050) | 11670..12002 | - | 333 | WP_000699980 | hypothetical protein | - |
| L1S38_RS24055 (L1S38_24055) | 12005..12400 | - | 396 | WP_000733627 | cag pathogenicity island Cag12 family protein | - |
| L1S38_RS24060 (L1S38_24060) | 12397..14232 | - | 1836 | WP_250670776 | type IV secretory system conjugative DNA transfer family protein | - |
| L1S38_RS24065 (L1S38_24065) | 14237..15274 | - | 1038 | WP_000217791 | P-type DNA transfer ATPase VirB11 | virB11 |
| L1S38_RS24070 (L1S38_24070) | 15292..16506 | - | 1215 | WP_025755765 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| L1S38_RS24075 (L1S38_24075) | 16503..17432 | - | 930 | WP_025755764 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| L1S38_RS24080 (L1S38_24080) | 17438..18160 | - | 723 | WP_000394570 | type IV secretion system protein | virB8 |
| L1S38_RS24085 (L1S38_24085) | 18350..19405 | - | 1056 | WP_000235774 | type IV secretion system protein | virB6 |
| L1S38_RS24090 (L1S38_24090) | 19417..19677 | - | 261 | WP_001228869 | EexN family lipoprotein | - |
| L1S38_RS24095 (L1S38_24095) | 19687..20424 | - | 738 | WP_000737859 | type IV secretion system protein | - |
| L1S38_RS24100 (L1S38_24100) | 20426..23185 | - | 2760 | WP_000108725 | VirB3 family type IV secretion system protein | virb4 |
| L1S38_RS24105 (L1S38_24105) | 23209..23499 | - | 291 | WP_000921916 | TrbC/VirB2 family protein | virB2 |
| L1S38_RS24110 (L1S38_24110) | 23483..24127 | - | 645 | WP_000953539 | lytic transglycosylase domain-containing protein | virB1 |
| L1S38_RS24115 (L1S38_24115) | 24352..24843 | - | 492 | WP_000872475 | transcription termination/antitermination NusG family protein | - |
| L1S38_RS24120 (L1S38_24120) | 25204..26361 | - | 1158 | WP_000538023 | DNA distortion polypeptide 2 | - |
| L1S38_RS24125 (L1S38_24125) | 26364..26909 | - | 546 | WP_025755763 | DNA distortion polypeptide 1 | - |
| L1S38_RS24130 (L1S38_24130) | 27249..27506 | - | 258 | WP_250670775 | hypothetical protein | - |
| L1S38_RS24135 (L1S38_24135) | 27522..27671 | - | 150 | WP_165839134 | hypothetical protein | - |
| L1S38_RS24140 (L1S38_24140) | 27748..28092 | - | 345 | WP_015058251 | hypothetical protein | - |
| L1S38_RS24145 (L1S38_24145) | 28188..28406 | - | 219 | WP_015058250 | hypothetical protein | - |
| L1S38_RS24150 (L1S38_24150) | 28399..28662 | - | 264 | WP_025755761 | hypothetical protein | - |
Host bacterium
| ID | 10547 | GenBank | NZ_CP091083 |
| Plasmid name | pHN21SC1505-2 | Incompatibility group | IncX2 |
| Plasmid size | 40695 bp | Coordinate of oriT [Strand] | 26681..27321 [+] |
| Host baterium | Enterobacter roggenkampii strain GD21SC1505 |
Cargo genes
| Drug resistance gene | qnrS1, tet(A) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |