Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 110063 |
| Name | oriT_kcgeb_e1|unnamed1 |
| Organism | Leclercia adecarboxylata strain kcgeb_e1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP098326 (24151..24250 [+], 100 nt) |
| oriT length | 100 nt |
| IRs (inverted repeats) | 79..84, 90..95 (AAAAAA..TTTTTT) 33..40, 43..50 (AGCGTGAT..ATCACGCT) 19..25, 37..43 (TAAATCA..TGATTTA) |
| Location of nic site | 61..62 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 100 nt
>oriT_kcgeb_e1|unnamed1
ATTTCTCTTTTTTGGTTTTAAATCATTGGGATAGCGTGATTTATCACGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
ATTTCTCTTTTTTGGTTTTAAATCATTGGGATAGCGTGATTTATCACGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file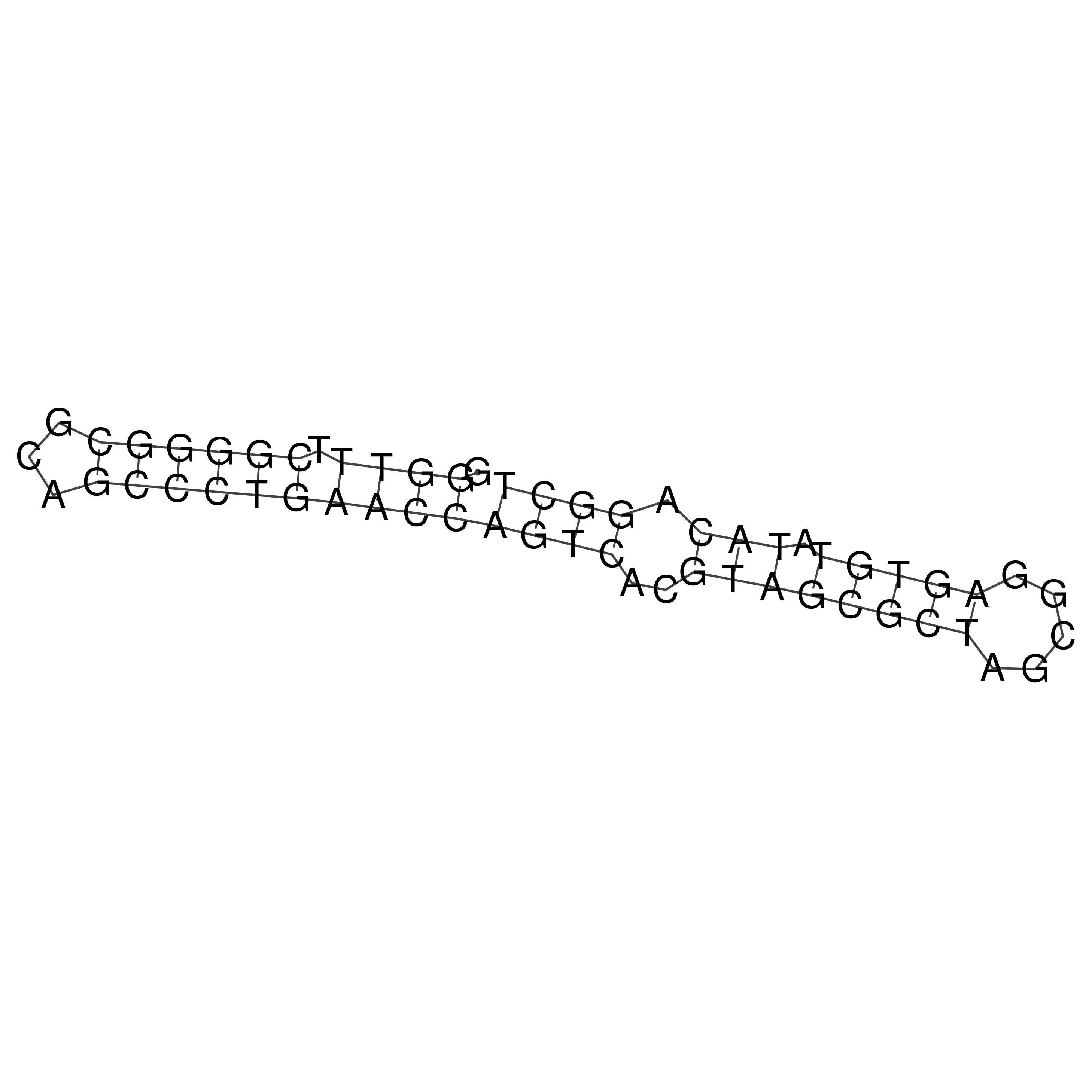
Relaxase
| ID | 6643 | GenBank | WP_129753792 |
| Name | mobF_NB069_RS22335_kcgeb_e1|unnamed1 |
UniProt ID | _ |
| Length | 1078 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1078 a.a. Molecular weight: 120075.28 Da Isoelectric Point: 6.7494
>WP_129753792.1 MULTISPECIES: MobF family relaxase [Enterobacteriaceae]
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTQMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTHKGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAEMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPHEADIARNTAPDFTSPQVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHASMVDVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDEGIKNGRLKKTSHRVTTVEGIRLERTILTIESRGRGQMPRLLTAEIAGQLLAGKTLKQEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGQKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAKLADRYLALSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGATFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYRVLDTGPGNKLTVES
SSGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKSQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSVEKGLTGVTGESMAFNQKPDEHNMTSGSDYQPVNNA
EDAFHLKQNPMDDSVGLRRHEAHHNDAELAHDYAAADDQQWSEQDYADYEHYAEAAEYDFDSSIYEDYAM
PQASQAEQSHAGKGHNHEHEHEEGGHEI
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTQMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTHKGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAEMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPHEADIARNTAPDFTSPQVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHASMVDVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDEGIKNGRLKKTSHRVTTVEGIRLERTILTIESRGRGQMPRLLTAEIAGQLLAGKTLKQEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGQKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAKLADRYLALSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGATFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYRVLDTGPGNKLTVES
SSGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKSQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSVEKGLTGVTGESMAFNQKPDEHNMTSGSDYQPVNNA
EDAFHLKQNPMDDSVGLRRHEAHHNDAELAHDYAAADDQQWSEQDYADYEHYAEAAEYDFDSSIYEDYAM
PQASQAEQSHAGKGHNHEHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 3585 | GenBank | WP_115719256 |
| Name | WP_115719256_kcgeb_e1|unnamed1 |
UniProt ID | _ |
| Length | 138 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 138 a.a. Molecular weight: 15380.56 Da Isoelectric Point: 5.9716
>WP_115719256.1 MULTISPECIES: traK protein [Enterobacteriaceae]
MPTITAKVSDELLAYIDRVSGGNRSEYLRRCLEAGPGDRESGLKIVADQLGDVNRKLDYLFDRASDADFG
SLRDELKAITETLSSVKFPPAGQMMLHESLAIETLILLRSIAEPGKNKAAKAELERNGYKIWEPKKER
MPTITAKVSDELLAYIDRVSGGNRSEYLRRCLEAGPGDRESGLKIVADQLGDVNRKLDYLFDRASDADFG
SLRDELKAITETLSSVKFPPAGQMMLHESLAIETLILLRSIAEPGKNKAAKAELERNGYKIWEPKKER
Protein domains
No domain identified.
Protein structure
No available structure.
T4CP
| ID | 7350 | GenBank | WP_115719258 |
| Name | t4cp2_NB069_RS22340_kcgeb_e1|unnamed1 |
UniProt ID | _ |
| Length | 509 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 509 a.a. Molecular weight: 57645.84 Da Isoelectric Point: 9.8124
>WP_115719258.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacteriaceae]
MDERERGLAFLFAITLPPVLVWFLVAKFTYGIDPSTAKYLVPYLVKNTFSLWPLWSALIAGWVIGIAGLV
AFILYDKTRVFKGERFKKIFRGTELVRARTLADKTRQRGVDQLTVANIPIPVEAENLHFSIAGTTGTGKT
TIFNELLFKSIKRGGKNIVLDPNGGFLKNFYRPGDAILNAYDKRTKGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYNTVTMEEVIHWACNVDQKKLKEFLTGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKKSLNPLISCWLDSIFSIVLGMGEKEGR
INVFIDELESLQYLPNLNDALTKGRKSGLCVFAGYQTFSQLVKVYGRDMAQTILANLRSNIVLGGSRLGE
DTLDHMSRSLGEIEGEVERKESDPQKPWIIRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KYIKYQRKNPVPGIELREI
MDERERGLAFLFAITLPPVLVWFLVAKFTYGIDPSTAKYLVPYLVKNTFSLWPLWSALIAGWVIGIAGLV
AFILYDKTRVFKGERFKKIFRGTELVRARTLADKTRQRGVDQLTVANIPIPVEAENLHFSIAGTTGTGKT
TIFNELLFKSIKRGGKNIVLDPNGGFLKNFYRPGDAILNAYDKRTKGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYNTVTMEEVIHWACNVDQKKLKEFLTGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKKSLNPLISCWLDSIFSIVLGMGEKEGR
INVFIDELESLQYLPNLNDALTKGRKSGLCVFAGYQTFSQLVKVYGRDMAQTILANLRSNIVLGGSRLGE
DTLDHMSRSLGEIEGEVERKESDPQKPWIIRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KYIKYQRKNPVPGIELREI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6591..23582
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NB069_RS22200 (NB069_22200) | 2364..2609 | + | 246 | WP_250589577 | AbrB/MazE/SpoVT family DNA-binding domain-containing protein | - |
| NB069_RS22205 (NB069_22205) | 2606..2980 | + | 375 | WP_250589578 | hypothetical protein | - |
| NB069_RS22210 (NB069_22210) | 2990..3316 | + | 327 | WP_250589579 | endoribonuclease MazF | - |
| NB069_RS22215 (NB069_22215) | 3318..3656 | + | 339 | WP_250589580 | hypothetical protein | - |
| NB069_RS22220 (NB069_22220) | 3748..3999 | + | 252 | WP_250589581 | hypothetical protein | - |
| NB069_RS22225 (NB069_22225) | 4017..4247 | + | 231 | Protein_7 | hypothetical protein | - |
| NB069_RS22230 (NB069_22230) | 4527..5099 | - | 573 | WP_250589582 | restriction endonuclease | - |
| NB069_RS22235 (NB069_22235) | 5149..5460 | - | 312 | WP_113458006 | hypothetical protein | - |
| NB069_RS22240 (NB069_22240) | 5506..5820 | - | 315 | WP_113458007 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| NB069_RS22245 (NB069_22245) | 5817..6161 | - | 345 | WP_250589583 | hypothetical protein | - |
| NB069_RS22250 (NB069_22250) | 6177..6482 | - | 306 | WP_249541702 | H-NS family nucleoid-associated regulatory protein | - |
| NB069_RS22255 (NB069_22255) | 6591..7325 | + | 735 | WP_033558688 | lytic transglycosylase domain-containing protein | virB1 |
| NB069_RS22260 (NB069_22260) | 7334..7615 | + | 282 | WP_032734186 | transcriptional repressor KorA | - |
| NB069_RS22265 (NB069_22265) | 7625..7918 | + | 294 | WP_033558690 | TrbC/VirB2 family protein | virB2 |
| NB069_RS22270 (NB069_22270) | 7971..8288 | + | 318 | WP_115719271 | VirB3 family type IV secretion system protein | virB3 |
| NB069_RS22275 (NB069_22275) | 8288..10888 | + | 2601 | WP_129753800 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| NB069_RS22280 (NB069_22280) | 10906..11619 | + | 714 | WP_250589584 | type IV secretion system protein | virB5 |
| NB069_RS22285 (NB069_22285) | 11627..11854 | + | 228 | WP_250589585 | IncN-type entry exclusion lipoprotein EexN | - |
| NB069_RS22290 (NB069_22290) | 11870..12919 | + | 1050 | WP_113458009 | type IV secretion system protein | virB6 |
| NB069_RS22590 | 13020..13148 | + | 129 | WP_071882546 | conjugal transfer protein TraN | - |
| NB069_RS22295 (NB069_22295) | 13138..13836 | + | 699 | WP_033558695 | type IV secretion system protein | virB8 |
| NB069_RS22300 (NB069_22300) | 13847..14731 | + | 885 | WP_033558696 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| NB069_RS22305 (NB069_22305) | 14731..15891 | + | 1161 | WP_033558697 | type IV secretion system protein VirB10 | virB10 |
| NB069_RS22310 (NB069_22310) | 15934..16929 | + | 996 | WP_250589586 | P-type DNA transfer ATPase VirB11 | virB11 |
| NB069_RS22315 (NB069_22315) | 16929..17462 | + | 534 | WP_113458011 | phospholipase D family protein | - |
| NB069_RS22320 (NB069_22320) | 17520..17762 | + | 243 | WP_250589587 | hypothetical protein | - |
| NB069_RS22325 (NB069_22325) | 17949..18179 | - | 231 | WP_129753790 | type II toxin-antitoxin system VapB family antitoxin | - |
| NB069_RS22330 (NB069_22330) | 18197..18817 | - | 621 | WP_250589588 | DUF6710 family protein | - |
| NB069_RS22335 (NB069_22335) | 18817..22053 | - | 3237 | WP_129753792 | MobF family relaxase | - |
| NB069_RS22340 (NB069_22340) | 22053..23582 | - | 1530 | WP_115719258 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| NB069_RS22345 (NB069_22345) | 23584..24000 | - | 417 | WP_115719256 | traK protein | - |
| NB069_RS22350 (NB069_22350) | 24525..24959 | + | 435 | WP_129753794 | plasmid stabilization protein StbA | - |
| NB069_RS22355 (NB069_22355) | 24968..25684 | + | 717 | WP_032734204 | StbB family protein | - |
| NB069_RS22360 (NB069_22360) | 25686..26054 | + | 369 | WP_113458019 | plasmid stabilization protein StbC | - |
| NB069_RS22365 (NB069_22365) | 26254..26598 | + | 345 | WP_250589567 | hypothetical protein | - |
| NB069_RS22370 (NB069_22370) | 26711..27028 | - | 318 | WP_250589568 | CcgAII protein | - |
| NB069_RS22375 (NB069_22375) | 27083..27262 | - | 180 | WP_115719248 | cobalamin biosynthesis protein CbiX | - |
Host bacterium
| ID | 10498 | GenBank | NZ_CP098326 |
| Plasmid name | kcgeb_e1|unnamed1 | Incompatibility group | - |
| Plasmid size | 42288 bp | Coordinate of oriT [Strand] | 24151..24250 [+] |
| Host baterium | Leclercia adecarboxylata strain kcgeb_e1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |