Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 109983 |
| Name | oriT_pHL21A |
| Organism | Salmonella enterica subsp. enterica strain HL21 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP126326 (28704..28765 [-], 62 nt) |
| oriT length | 62 nt |
| IRs (inverted repeats) | 18..25, 28..35 (GCAAAATT..AATTTTGC) |
| Location of nic site | 44..45 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 62 nt
TTTATAAATAAAAATCAGCAAAATTTTAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file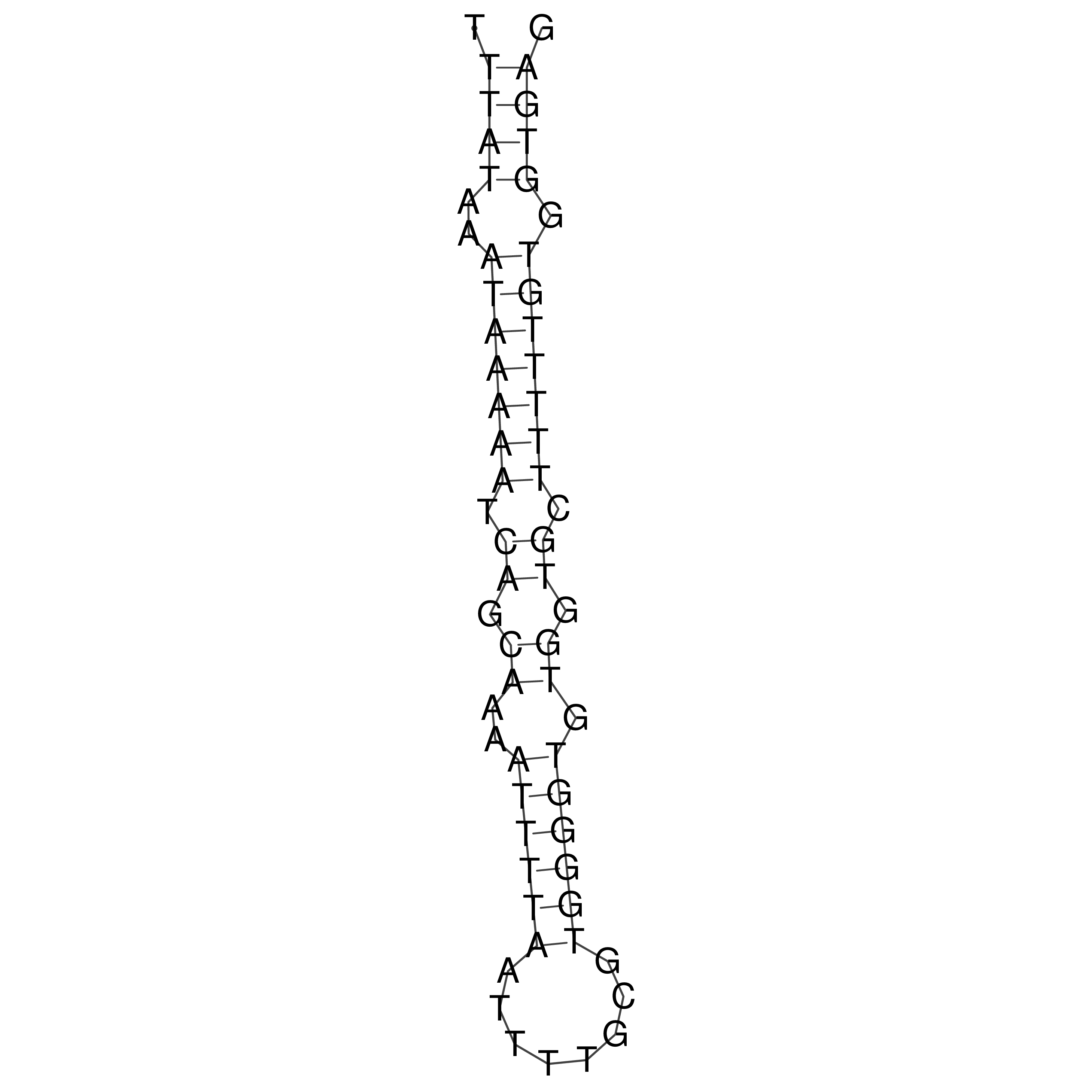
Relaxase
| ID | 6591 | GenBank | WP_079975180 |
| Name | traI_QN088_RS23725_pHL21A |
UniProt ID | _ |
| Length | 1755 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1755 a.a. Molecular weight: 191781.65 Da Isoelectric Point: 5.7503
MMSIAQVRSAGSAAGYYSDRDNYYVLGSLEERWAGKGAEQLGLQGAVDKEVFTRVLEGRLPDGADLSRQQ
DGGNKHRPGYDLTFSAPKSVSLMAMLAGDKRLTEAHNQAVDIAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTLHDGEWKTLSSDKVGKTGFIENVYANQIAFGKIYRAVLKEKVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQTIREAVGDDASLKSRDVAALDTRKSKQHVDPEVKMAEWMQT
LKDTGFDISAYREAADRRAEIQAAQPVPSQEQPDIQQAVTQAIAGLSDRKVQFTYTDVLARTVGMLPPEA
GVIEKARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSSDIMKQNRVTVYPEKSVPRTGSYSD
AVSVLAQDRPSLAIISGQGGAAGQRERVAELTMMAREQGREVQIIAADRRSQTNLKQDERLSGELITGRR
QLQEGMSFSPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKEAGVNSYRWQ
GGQQTPATVISEPDRNVRYARLAGDFVAAVKAGEESVAQVSGVREQAILAGMIRSELKTQGILGQQDTMM
TALSPVWLDSRNRYLRDMYREGMVMEQWNPEKRSHDRYVIDRVTAQSHSLTLRDAQGETQMVRISALDSS
WSLFRPEKIPVADGERLMVTGKIPGLRVSGGDRLQVSAVNDGMMTVIVPGRAEPASLPVGDSPFTALKLE
NGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLSRHPSFTVVSEQIK
ARAGEVSLETAISRQKAGLHTPAQQAIHLALPVVESKNLAFSQVDLLTEAKSFAAEGTGFADLGREIDAQ
IKRGDLLHVDVAKGYGTDLLISRASYDAEKSILHHILEGKEAVTPLMERVPGELMETLTSGQRAATRMIL
ETKDRFTVVQGYAGVGKTTQFRAVMSAVNLLPESERPRVVGLGPTHRAVGEMRSAGVEAQTLASFLHDTQ
LQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSAA
DVAIMKEIVRQTPELRDAVYSLINRDVNKALSGLENVKPVQVPRLKGAWAPENSVTEFSRLQERELAKAA
QEAEKKGEAFPDVPVTLHEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDVREKAGELGEQQADI
PVLTTANIRDGELRRLSTWEAHQGALALVDRVYHRIAGISKEDGLITLEDKEGNIRLISPREASAEGVTL
YKPETIRVGTGDRMRFTKSDPERGYVANSVWTVTAVSGDSVTLSDGKQTRVVRPGQDRAEQHIDLAYAIT
AHGAQGASETFAIALEGTEGGRKQMAGFESAYVALSRMKQHVQVYTDDRQGWVKAINSAEQKGTAHDVLE
PKSEREMMNAERLFSTARELRDVAAGRAVLRNAGLAQGDSRARFIAPGRKYPQPYVALPAFDRNGKSAGI
WLNPLTTDDGAGLRGFTGEGRVKGSEEAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGDG
RPWNPGAITGGRVWGDIPDSSVQPGAGNGEPVTAEILAQQQAEEAVRRETEQRAAEIVRKMAEDKTDLPE
EKTAQTVREVAGQEQDRMTPPERETPLPESVLREPVRERETIREVARENRVRERLQQMEQEMVRDLQKEK
TPGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 3542 | GenBank | WP_001151568 |
| Name | WP_001151568_pHL21A |
UniProt ID | Q8L369 |
| Length | 126 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 126 a.a. Molecular weight: 14293.25 Da Isoelectric Point: 4.8463
MAKVQAYVSDEVVEKINAIVEKRRSEGAKSTDVSFSSISTMLLELGLRVYEAQMERKESAFNQMEFNRVL
LENVLKTQSSVVKILGIGSISPHVAGNPKFEYANMVEDIKEKVSSEMERFFHENDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q8L369 |
| ID | 3543 | GenBank | WP_227005938 |
| Name | WP_227005938_pHL21A |
UniProt ID | _ |
| Length | 133 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 133 a.a. Molecular weight: 15471.78 Da Isoelectric Point: 10.5644
MSLKRFGTRSATGKMVKLKLPVDVEKLLREASDRSGRSRSFEAVIRLKDHLHRYPKFNRKGNVYGKSLVK
YLTMRLDDETNQLLITAKNRSGWCKTDEAADRVIDHLTKFPDFYNSDIFREANEGKKSTFKEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 7278 | GenBank | WP_284546342 |
| Name | traC_QN088_RS23625_pHL21A |
UniProt ID | _ |
| Length | 882 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 882 a.a. Molecular weight: 100531.27 Da Isoelectric Point: 6.6546
MKNLLDTVTQAANSLLTALKLPDESAQANQILGEMSFPQFSRLLPYRDYNQESGLFMNDATMGFMLEATP
VNGANETIALSLDHLLRTKLPRGIPLCVHLMSSQLVGERIEYGLREFSWSGEQAERFNAITRAYYMKAAE
THFPLPEGMNLPLTLRHYRVFISYCAPSRKKSRADILEMENLVKIIRASLQGAYISTQTVDAQAFINIVG
EMINHNPESLHPTRRRLDPYSDLNYQCVDDSFDLKVRQDYLTLGLRDHGKNSTARIMNFHLARNPEIAFL
WNSADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGD
LRQRLASNQSSVVSYFFNITAFCRDNKETALETEQDILNSFRKNGFELISPRFNHMRNFLACLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERIRDQLSV
MASPNGNLDEVHEGLLLQAVRAAWLTKKNRARIDDVVDFLENARTSEQYAESPGIRSRLDEMIVLLNQYT
AAGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNRKVGDFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFPQLHREMIEKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLQEHETFRRTYEYRAETD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 7279 | GenBank | WP_284546308 |
| Name | traD_QN088_RS23710_pHL21A |
UniProt ID | _ |
| Length | 756 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 756 a.a. Molecular weight: 85739.61 Da Isoelectric Point: 5.2473
MSFNAKDMTQGGQIASMRIRMFSQIANIIFYCLFIFFWILVGLVLWVKLSWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIRYYGQTFRMNAAQVLQDKYTVWCGEQLWSAFVLAACVALVICLITFFMVTWILGRQGKQ
QSEDDVTGGRQLTDNPKDVARMLKKDGKASDILIGDLPIIKDSEIQNFLLHGTVGAGKSEIIRRLANYAR
QRGDMVVMYDRSGEFVKSYYDPSIDKILNPRDARCAAWDLWRECLTLPDFDNAANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDRDRSYSKLVDTLLSIKIEKLRTYLRDSSAANLVEEKIEKTAISIRAVLTNYVKA
VRYLQGIERNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEIVPEARKFGGCYVFGIQSSTQMEDIYGEKAAATLLDVMNTRAFFRSPSYKIA
DYVAHEIGEKEILKASEQYSYGADPVRDGVSTGKEMERVTLVSYSDIQSLPDLTCYVTLPGPYPAVKLTL
KYQERPKVAPEFIPREMNPEAEARLSAVLAAREAEGRQMSSLFEPDTEAVAPTAAEAQAEQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQPQQPQQPQQPQPTVVSDKKSAAASTAVNTPAGGVGQELKMKPEDEEQPL
PPGINASGEVVDMAAYERWRQEQEVNTQQQMQRREEVNINVHRGHGKDEPEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 28160..55744
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QN088_RS23525 (QN088_23525) | 23315..25312 | + | 1998 | WP_284546340 | ParB/RepB/Spo0J family partition protein | - |
| QN088_RS23530 (QN088_23530) | 25352..25786 | + | 435 | WP_079975213 | conjugation system SOS inhibitor PsiB | - |
| QN088_RS23535 (QN088_23535) | 25783..26520 | + | 738 | WP_284546341 | plasmid SOS inhibition protein A | - |
| QN088_RS23540 (QN088_23540) | 26769..26927 | + | 159 | WP_176452916 | hypothetical protein | - |
| QN088_RS23545 (QN088_23545) | 27030..27849 | + | 820 | Protein_38 | DUF932 domain-containing protein | - |
| QN088_RS23550 (QN088_23550) | 28160..28630 | - | 471 | WP_042828200 | transglycosylase SLT domain-containing protein | virB1 |
| QN088_RS23555 (QN088_23555) | 29054..29434 | + | 381 | WP_001151568 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| QN088_RS23560 (QN088_23560) | 29629..30318 | + | 690 | WP_079841988 | conjugal transfer transcriptional regulator TraJ | - |
| QN088_RS23565 (QN088_23565) | 30417..30812 | + | 396 | WP_001674566 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| QN088_RS23570 (QN088_23570) | 30854..31216 | + | 363 | WP_001274202 | type IV conjugative transfer system pilin TraA | - |
| QN088_RS23575 (QN088_23575) | 31231..31542 | + | 312 | WP_000012129 | type IV conjugative transfer system protein TraL | traL |
| QN088_RS23580 (QN088_23580) | 31564..32130 | + | 567 | WP_079975214 | type IV conjugative transfer system protein TraE | traE |
| QN088_RS23585 (QN088_23585) | 32117..32857 | + | 741 | WP_079955193 | type-F conjugative transfer system secretin TraK | traK |
| QN088_RS23590 (QN088_23590) | 32857..34278 | + | 1422 | WP_079975215 | F-type conjugal transfer pilus assembly protein TraB | traB |
| QN088_RS23595 (QN088_23595) | 34268..34864 | + | 597 | WP_058108410 | conjugal transfer pilus-stabilizing protein TraP | - |
| QN088_RS23600 (QN088_23600) | 34842..35087 | + | 246 | WP_077950176 | DUF2689 domain-containing protein | virb4 |
| QN088_RS23605 (QN088_23605) | 35084..35599 | + | 516 | WP_079975216 | type IV conjugative transfer system lipoprotein TraV | traV |
| QN088_RS23610 (QN088_23610) | 35734..35955 | + | 222 | WP_001278699 | conjugal transfer protein TraR | - |
| QN088_RS23615 (QN088_23615) | 35948..36415 | + | 468 | WP_079955208 | hypothetical protein | - |
| QN088_RS23620 (QN088_23620) | 36412..36795 | + | 384 | WP_079955209 | hypothetical protein | - |
| QN088_RS23625 (QN088_23625) | 36811..39459 | + | 2649 | WP_284546342 | type IV secretion system protein TraC | virb4 |
| QN088_RS23630 (QN088_23630) | 39434..39820 | + | 387 | WP_053253594 | type-F conjugative transfer system protein TrbI | - |
| QN088_RS23635 (QN088_23635) | 39817..40449 | + | 633 | WP_079975218 | type-F conjugative transfer system protein TraW | traW |
| QN088_RS23640 (QN088_23640) | 40446..41438 | + | 993 | WP_079975219 | conjugal transfer pilus assembly protein TraU | traU |
| QN088_RS23645 (QN088_23645) | 41457..41765 | + | 309 | WP_079975220 | hypothetical protein | - |
| QN088_RS23650 (QN088_23650) | 41750..42388 | + | 639 | WP_079975221 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| QN088_RS23655 (QN088_23655) | 42425..42844 | + | 420 | WP_021003392 | HNH endonuclease signature motif containing protein | - |
| QN088_RS23660 (QN088_23660) | 42841..44667 | + | 1827 | WP_079975222 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| QN088_RS23665 (QN088_23665) | 44678..44893 | + | 216 | WP_000700023 | conjugal transfer protein TrbE | - |
| QN088_RS23670 (QN088_23670) | 44905..45654 | + | 750 | WP_079975223 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| QN088_RS23675 (QN088_23675) | 45868..46161 | + | 294 | WP_001233881 | type-F conjugative transfer system pilin chaperone TraQ | - |
| QN088_RS23680 (QN088_23680) | 46148..46696 | + | 549 | WP_079955199 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| QN088_RS23685 (QN088_23685) | 46686..48068 | + | 1383 | WP_001161720 | conjugal transfer pilus assembly protein TraH | traH |
| QN088_RS23690 (QN088_23690) | 48065..50887 | + | 2823 | WP_079975225 | conjugal transfer mating-pair stabilization protein TraG | traG |
| QN088_RS23695 (QN088_23695) | 50909..51388 | + | 480 | WP_284546307 | surface exclusion protein | - |
| QN088_RS23700 (QN088_23700) | 51734..52465 | + | 732 | WP_079975227 | conjugal transfer complement resistance protein TraT | - |
| QN088_RS23705 (QN088_23705) | 52717..53196 | + | 480 | WP_079975228 | hypothetical protein | - |
| QN088_RS23710 (QN088_23710) | 53474..55744 | + | 2271 | WP_284546308 | type IV conjugative transfer system coupling protein TraD | virb4 |
| QN088_RS23715 (QN088_23715) | 55753..56151 | - | 399 | WP_000911322 | type II toxin-antitoxin system VapC family toxin | - |
| QN088_RS23720 (QN088_23720) | 56151..56378 | - | 228 | WP_000450263 | toxin-antitoxin system antitoxin VapB | - |
Host bacterium
| ID | 10418 | GenBank | NZ_CP126326 |
| Plasmid name | pHL21A | Incompatibility group | - |
| Plasmid size | 90249 bp | Coordinate of oriT [Strand] | 28704..28765 [-] |
| Host baterium | Salmonella enterica subsp. enterica strain HL21 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |