Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 109798 |
| Name | oriT_C. koseri 0|P1 |
| Organism | Citrobacter koseri isolate 0 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_OW969692 (26794..26866 [-], 73 nt) |
| oriT length | 73 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 73 nt
>oriT_C. koseri 0|P1
GTCGGGGCAAAGCCCTGACCAGGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
GTCGGGGCAAAGCCCTGACCAGGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file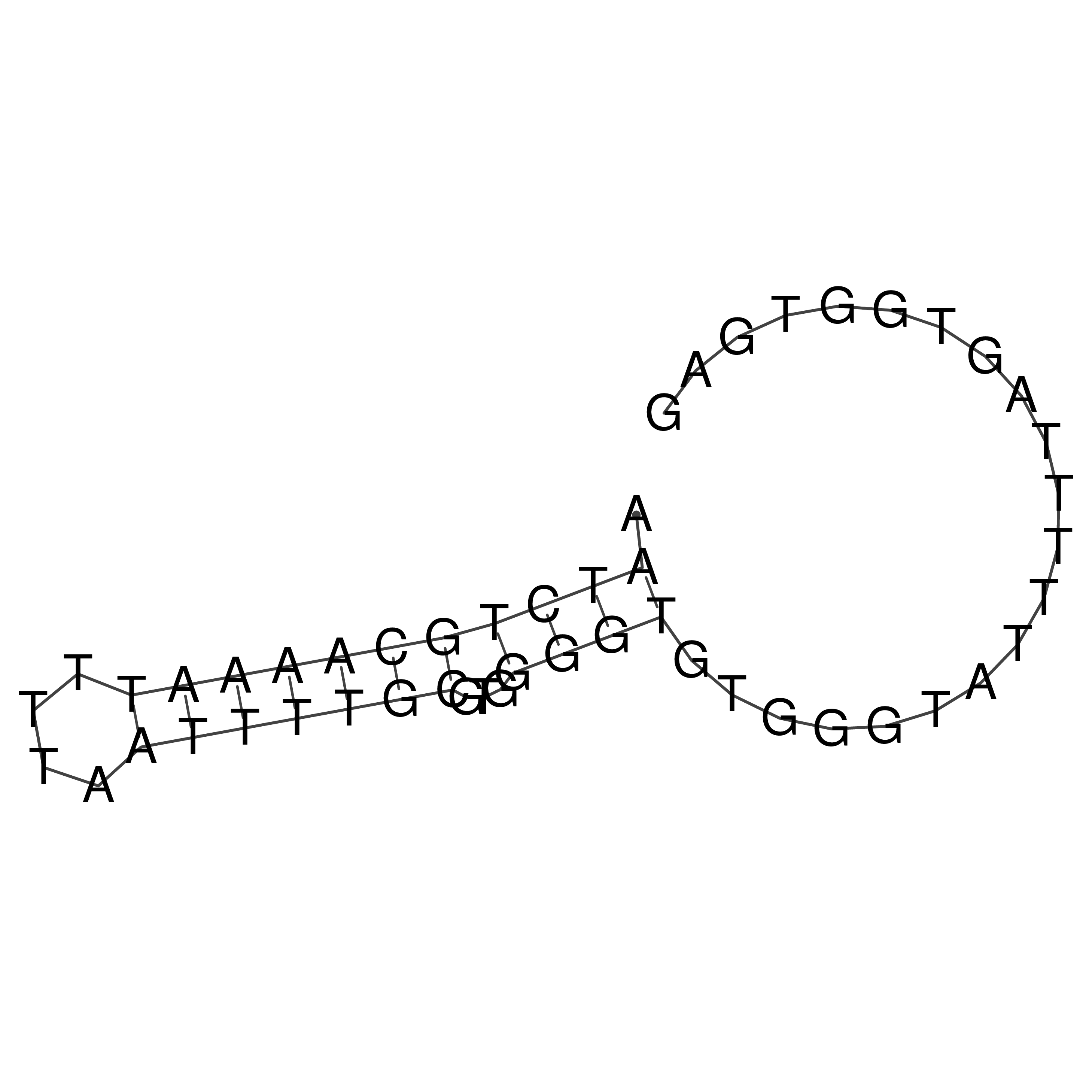
Relaxase
| ID | 6483 | GenBank | WP_000991384 |
| Name | Relaxase_LQV22_RS22315_C. koseri 0|P1 |
UniProt ID | A0A8H8Z9E5 |
| Length | 903 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 903 a.a. Molecular weight: 104595.99 Da Isoelectric Point: 7.6377
>WP_000991384.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPEMARNGELESCLDVHRTLAKAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGRNLGTQAVSDYEWYRIRMGTGPQGAIKRELFSDKESLWGY
TTVQCESLIEDMIAGGNFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAGADIFERVKPECRYNPELAASDEVELGFRRDPVLRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAVQGDRAALSQLRGWDYRDRRKDKRRTTNADRCVILCEPGGTPLYEDTGVLEARLQKDGS
VRFRDRRNGELVCVDYGDRVVFYHHQDRNELVDKLNLIAPVLFDREPRMGFEPEGSYQQFNDVFAEMVAW
HNAAGITGSGHFVISRPDVDLHRQRSEQYYHEYIRQQKSISGGHGASYAPVQDNEWTPPSPGM
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPEMARNGELESCLDVHRTLAKAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGRNLGTQAVSDYEWYRIRMGTGPQGAIKRELFSDKESLWGY
TTVQCESLIEDMIAGGNFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAGADIFERVKPECRYNPELAASDEVELGFRRDPVLRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAVQGDRAALSQLRGWDYRDRRKDKRRTTNADRCVILCEPGGTPLYEDTGVLEARLQKDGS
VRFRDRRNGELVCVDYGDRVVFYHHQDRNELVDKLNLIAPVLFDREPRMGFEPEGSYQQFNDVFAEMVAW
HNAAGITGSGHFVISRPDVDLHRQRSEQYYHEYIRQQKSISGGHGASYAPVQDNEWTPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 3421 | GenBank | WP_001291058 |
| Name | WP_001291058_C. koseri 0|P1 |
UniProt ID | A0A7I0L241 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12730.52 Da Isoelectric Point: 9.9656
>WP_001291058.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7I0L241 |
T4CP
| ID | 7106 | GenBank | WP_000077663 |
| Name | trbC_LQV22_RS22400_C. koseri 0|P1 |
UniProt ID | _ |
| Length | 767 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 767 a.a. Molecular weight: 87012.13 Da Isoelectric Point: 7.3008
>WP_000077663.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSQHHVNPDLIHRTAWGNPLWNALHNLNITGLCLAGSIITALIWPLALPVCLLFTLVTSVIFMLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETFPARGIFYVGYRRINDIGRELWLSMDDLTRHVM
FFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMLLARDLGYRLE
GTDAQTLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTAKPSRKRRKNRTEPYRIIDTFQRQLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEMLLPVPDLTASPPHKKNVAQTPSGGKQESRKKRF
MSQHHVNPDLIHRTAWGNPLWNALHNLNITGLCLAGSIITALIWPLALPVCLLFTLVTSVIFMLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETFPARGIFYVGYRRINDIGRELWLSMDDLTRHVM
FFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMLLARDLGYRLE
GTDAQTLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTAKPSRKRRKNRTEPYRIIDTFQRQLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEMLLPVPDLTASPPHKKNVAQTPSGGKQESRKKRF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 48911..92906
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LQV22_RS22385 | 43977..44084 | - | 108 | Protein_52 | aminoglycoside phosphotransferase family protein | - |
| LQV22_RS22390 | 44084..44887 | - | 804 | WP_001082319 | aminoglycoside O-phosphotransferase APH(3'')-Ib | - |
| LQV22_RS22395 (AN2351V1_4340) | 44948..45763 | - | 816 | WP_001043260 | sulfonamide-resistant dihydropteroate synthase Sul2 | - |
| LQV22_RS22400 (AN2351V1_4342) | 46627..48930 | - | 2304 | WP_000077663 | F-type conjugative transfer protein TrbC | - |
| LQV22_RS22405 (AN2351V1_4343) | 48911..50035 | - | 1125 | WP_001557490 | DsbC family protein | trbB |
| LQV22_RS22410 (AN2351V1_4344) | 50032..51321 | - | 1290 | WP_000121916 | IncI1-type conjugal transfer protein TrbA | trbA |
| LQV22_RS22415 | 51659..51805 | + | 147 | WP_001346191 | hypothetical protein | - |
| LQV22_RS23145 (AN2351V1_4345) | 51858..51953 | + | 96 | WP_001388983 | DinQ-like type I toxin DqlB | - |
| LQV22_RS23150 (AN2351V1_4346) | 52542..52637 | + | 96 | WP_001388983 | DinQ-like type I toxin DqlB | - |
| LQV22_RS22425 (AN2351V1_4347) | 52700..52876 | - | 177 | WP_001054907 | hypothetical protein | - |
| LQV22_RS22430 (AN2351V1_4348) | 53026..53493 | - | 468 | WP_000598962 | thermonuclease family protein | - |
| LQV22_RS22435 (AN2351V1_4349) | 53653..54276 | + | 624 | WP_001155250 | helix-turn-helix domain-containing protein | - |
| LQV22_RS22440 (AN2351V1_4351) | 54474..54689 | - | 216 | WP_000266543 | hypothetical protein | - |
| LQV22_RS22445 (AN2351V1_4352) | 54693..55061 | - | 369 | WP_001345852 | hypothetical protein | - |
| LQV22_RS22450 (AN2351V1_4353) | 55352..55594 | - | 243 | WP_000650923 | hypothetical protein | - |
| LQV22_RS22455 (AN2351V1_4354) | 55924..56076 | + | 153 | WP_001335079 | Hok/Gef family protein | - |
| LQV22_RS22460 (AN2351V1_4355) | 56214..57800 | - | 1587 | WP_000004564 | TnsA endonuclease N-terminal domain-containing protein | - |
| LQV22_RS22465 (AN2351V1_4356) | 58074..58721 | - | 648 | WP_015056434 | plasmid IncI1-type surface exclusion protein ExcA | - |
| LQV22_RS22470 (AN2351V1_4357) | 58809..60968 | - | 2160 | WP_000691811 | DotA/TraY family protein | traY |
| LQV22_RS22475 (AN2351V1_4358) | 61043..61612 | - | 570 | WP_000650752 | conjugal transfer protein TraX | - |
| LQV22_RS22480 (AN2351V1_4359) | 61609..62814 | - | 1206 | WP_001189548 | conjugal transfer protein TraW | traW |
| LQV22_RS22485 (AN2351V1_4360) | 62772..63392 | - | 621 | WP_000286855 | hypothetical protein | traV |
| LQV22_RS22490 (AN2351V1_4361) | 63392..66436 | - | 3045 | WP_001024769 | conjugal transfer protein | traU |
| LQV22_RS22495 (AN2351V1_4362) | 66572..66760 | - | 189 | Protein_75 | IS66 family transposase | - |
| LQV22_RS22500 (AN2351V1_4363) | 66821..67954 | + | 1134 | WP_000555378 | IS110-like element ISEc45 family transposase | - |
| LQV22_RS22505 (AN2351V1_4364) | 68207..69631 | - | 1425 | Protein_77 | IS66-like element ISEc23 family transposase | - |
| LQV22_RS22510 (AN2351V1_4365) | 69662..70012 | - | 351 | WP_000624722 | IS66 family insertion sequence element accessory protein TnpB | - |
| LQV22_RS22515 (AN2351V1_4366) | 70009..70434 | - | 426 | WP_000422741 | transposase | - |
| LQV22_RS22520 (AN2351V1_4367) | 70717..71343 | - | 627 | WP_000785703 | hypothetical protein | traT |
| LQV22_RS23155 (AN2351V1_4368) | 71450..71701 | - | 252 | WP_001277573 | hypothetical protein | - |
| LQV22_RS22530 (AN2351V1_4369) | 71758..72156 | - | 399 | WP_000088813 | DUF6750 family protein | traR |
| LQV22_RS22535 (AN2351V1_4370) | 72203..72733 | - | 531 | WP_000987021 | conjugal transfer protein TraQ | traQ |
| LQV22_RS22540 (AN2351V1_4371) | 72730..73443 | - | 714 | WP_000787002 | conjugal transfer protein TraP | traP |
| LQV22_RS22545 (AN2351V1_4372) | 73440..74777 | - | 1338 | WP_001557492 | conjugal transfer protein TraO | traO |
| LQV22_RS22550 (AN2351V1_4373) | 74781..75755 | - | 975 | WP_000547386 | DotH/IcmK family type IV secretion protein | traN |
| LQV22_RS22555 (AN2351V1_4374) | 75766..76461 | - | 696 | WP_001287545 | DotI/IcmL family type IV secretion protein | traM |
| LQV22_RS22560 (AN2351V1_4375) | 76473..76823 | - | 351 | WP_068862892 | conjugal transfer protein | traL |
| LQV22_RS22565 (AN2351V1_4376) | 76840..80901 | - | 4062 | WP_000842456 | LPD7 domain-containing protein | - |
| LQV22_RS22570 (AN2351V1_4377) | 80965..81255 | - | 291 | WP_000817801 | IcmT/TraK family protein | traK |
| LQV22_RS22575 (AN2351V1_4378) | 81252..82400 | - | 1149 | WP_000775210 | plasmid transfer ATPase TraJ | virB11 |
| LQV22_RS22580 (AN2351V1_4379) | 82384..83220 | - | 837 | WP_000745049 | type IV secretory system conjugative DNA transfer family protein | traI |
| LQV22_RS22585 (AN2351V1_4380) | 83217..83675 | - | 459 | WP_001170111 | DotD/TraH family lipoprotein | - |
| LQV22_RS22590 (AN2351V1_4381) | 83779..84981 | - | 1203 | WP_000979993 | conjugal transfer protein TraF | - |
| LQV22_RS22595 (AN2351V1_4382) | 85083..85904 | - | 822 | WP_000845373 | hypothetical protein | traE |
| LQV22_RS22600 | 86069..86263 | - | 195 | Protein_96 | integrase | - |
| LQV22_RS22605 (AN2351V1_REPA000000007) | 86382..87079 | + | 698 | WP_095033700 | IS1-like element IS1B family transposase | - |
| LQV22_RS22610 (AN2351V1_4386) | 87153..88514 | - | 1362 | WP_001168171 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| LQV22_RS22615 (AN2351V1_4387) | 88532..89158 | - | 627 | WP_000939245 | prepilin peptidase | - |
| LQV22_RS22620 (AN2351V1_4388) | 89174..89659 | - | 486 | WP_001080767 | lytic transglycosylase domain-containing protein | virB1 |
| LQV22_RS22625 (AN2351V1_4389) | 89704..90240 | - | 537 | WP_000111534 | type 4 pilus major pilin | - |
| LQV22_RS22630 (AN2351V1_4390) | 90302..91396 | - | 1095 | WP_000697158 | type II secretion system F family protein | - |
| LQV22_RS22635 (AN2351V1_4391) | 91398..92906 | - | 1509 | WP_000880805 | ATPase, T2SS/T4P/T4SS family | virB11 |
| LQV22_RS22640 (AN2351V1_4392) | 93010..93468 | - | 459 | WP_000095885 | type IV pilus biogenesis protein PilP | - |
| LQV22_RS22645 (AN2351V1_4393) | 93458..94753 | - | 1296 | WP_000129882 | type 4b pilus protein PilO2 | - |
| LQV22_RS22650 (AN2351V1_4394) | 94774..96393 | - | 1620 | WP_001035565 | PilN family type IVB pilus formation outer membrane protein | - |
| LQV22_RS22655 (AN2351V1_4395) | 96425..96862 | - | 438 | WP_000539952 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 10233 | GenBank | NZ_OW969692 |
| Plasmid name | C. koseri 0|P1 | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 103689 bp | Coordinate of oriT [Strand] | 26794..26866 [-] |
| Host baterium | Citrobacter koseri isolate 0 |
Cargo genes
| Drug resistance gene | floR, aph(6)-Id, blaTEM-1C, aph(3'')-Ib, sul2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |