Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108937 |
| Name | oriT_pB-4515_1 |
| Organism | Salmonella enterica subsp. enterica serovar Typhimurium strain SCPM-O-B-4515 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP088137 (53102..53163 [-], 62 nt) |
| oriT length | 62 nt |
| IRs (inverted repeats) | 18..25, 28..35 (GCAAAATT..AATTTTGC) |
| Location of nic site | 44..45 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 62 nt
TTTATAAATAAAAATCAGCAAAATTTTAATTTTGCGTGGGGTGTGGTGCTTTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file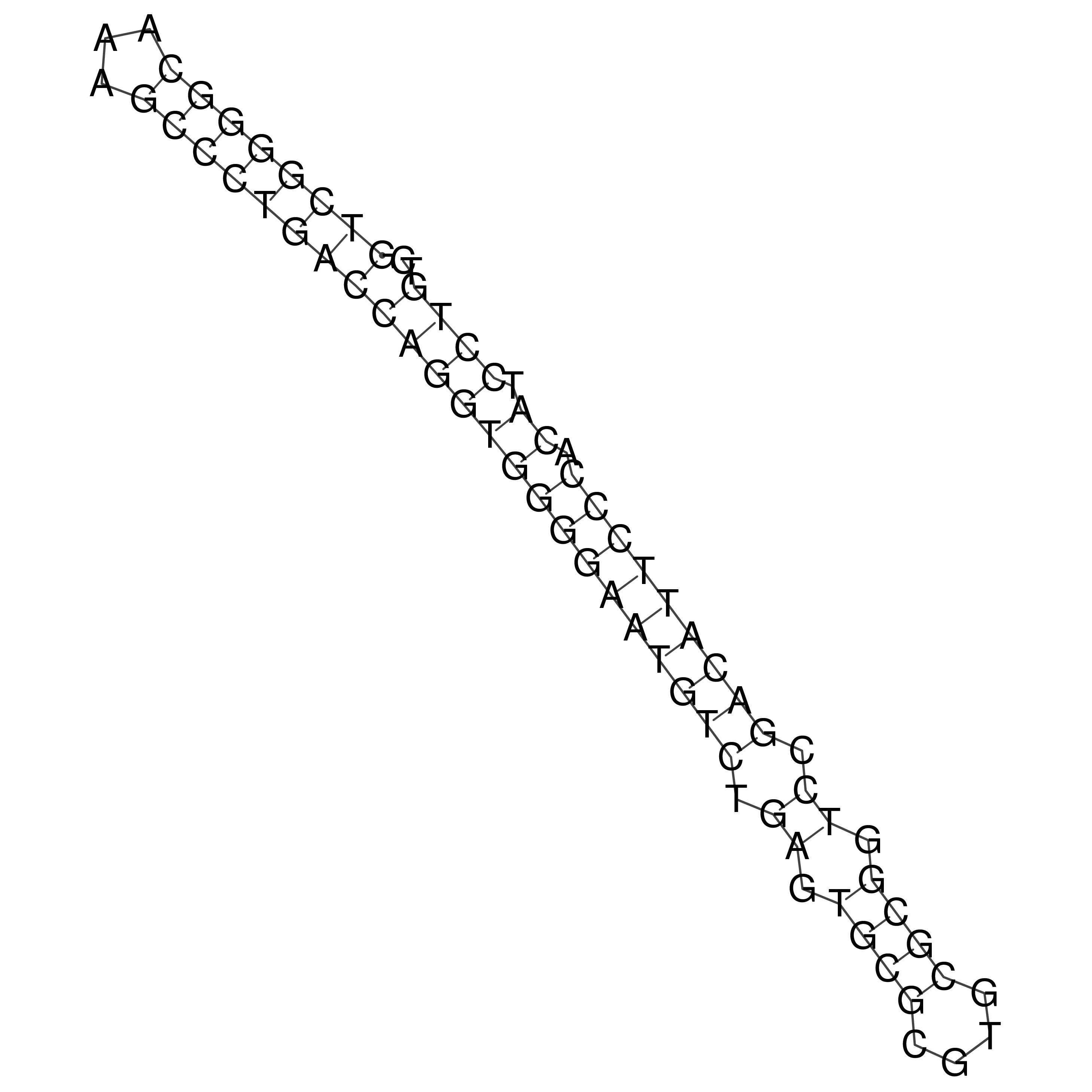
Relaxase
| ID | 5991 | GenBank | WP_249895813 |
| Name | traI_LRM91_RS22960_pB-4515_1 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 191327.24 Da Isoelectric Point: 5.9174
MMSIAQVRSAGSAAGYYSDRDNYYVLGSLEERWAGKGAEQLGVQGTVDKEVFTRVLEGRLPDGADLSRQQ
DGGNKHRPGYDLTFSAPKSVSLIAMLAGDKRLTEAHNQAVDIAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTLHDGEWKTLSSDKVGKTGFIENVYANQIAFGKIYRAVLKEKVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVKMAEWMQT
LKDTGFDISAYREAADRRAEIQAAQPVPSQEQPDIQQAVTQAIAGLSDRKVQFTYTDVLARTVGMLPPEA
GVIEKARAGIDEAISREWLIPLDREKGLFTSGIHVLDELSVRALSSDIMKQNRVTVHPEKSVPRTGSYSD
AVSVLAQDRPSLAIISGQGSAAGQRERVAELTMMAREQGREVQIIAADRRSQTNLKQDERLSGELITGRR
QLQEGMSFSPGSTVIVDQGEKLSLKETLTLLDGAARHNIQVLITDSGQRTGTGSALMAMKEAGVNSYRWQ
GGQQTPATVISGPDRNVRYARLAGDFVAAVKAGEESVAQVSGVREQAILAGMIRSELKTQGVLGQQDTMM
TALSPVWLDSRNRYLRDMYREGMVMEQWNPEKRSHDRYVIDRVTAQSHSLTLRDAQGETQMVRISALDSS
WSLFRPEKIPVADGERLMVTGKIPGLRVSGGDRLQVSAVNDGMMTVIVPGRAEPASLPVGDSPFTALKLE
NGWVETPGHSVSDSAKVFVSVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLSRHPSFTVVSEQIK
ARAGEVSLETAISRQKAGLHTPAQQAIHLALPVVESKNLAFSQVELLTEAKSFAAEGTGFAELGREIDAQ
IKRGDLLHVDVAKGYGTDLLISRASYDAEKSILHHILEGKEAVTPLMERVSGELMETLTSGQRAATRMIL
ETKDRFTVVQGYAGVGKTTQFRAVMSAVNLLPGSERPRVVGLGPTHRAVGEMRSAGVEAQTLASFLHDTQ
LQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSAA
DVAIMKEIVRQTPELRDAVYSLINRDVNKALSGLENVKPVQVPRLKGAWVPENSVTEFSRLQERELVKAA
QEAEKKGEAFPDVPVTLYEAIVRDYTGRTPDAREQTLIVTHLNEDRRVLNSMIQDALAKPGEQQVTVPVL
TTANIRDGELRRLSTWENHQCALALVDKVYHRIAGISKEDGLITLQDADGNTRLISPREAAAEGVTLYNP
ETIRVGAGDRMRFTKSDRERGYVANSVWPVTAVSGDGVTLSDGKQTRVVRPGQDRAEQHIDLAYAITAHG
AQGASETFAIALEGTEGGRKQMAGFESAYVALSRMKQHVQVYTDDRQGWTDAINRAAQKGTAHDVLEPKA
DREAMNAERLFSTARELRDVAAGRAVLRNAGLALGDSRARFIAPGRKYPQPYVALPAFDRNGKSAGIWLN
PLTTDDGAGLRGFSGEGRVKGSEEAQFVALQGSRNGESLLADNMQEGVRIARDNPDSGVVVRIAGDGRPW
NPGAITGGRVWGDIPDSSVQPGAVNGEPVTAEILAQRQAEEAVRRETEQRAAEIVRKMAEDKPDLPEEKT
AQAVREIAGQEQDRMTPPERETPLPESVLREPVRERETIREVARENRVRERLQQTEQEMVRDLQKERTPD
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 3019 | GenBank | WP_249895802 |
| Name | WP_249895802_pB-4515_1 |
UniProt ID | _ |
| Length | 126 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 126 a.a. Molecular weight: 14351.29 Da Isoelectric Point: 4.7257
MAKVQAYVSDEVVEKINAIVEKRRSEGEKSTDVSFSSISTMLLELGLRVYEAQMERKESAFNQMEFNRVL
LENVLKTQSSVVKILGIGSISPHVAGNPKFEYANMVEDIKEKVSSEMERFFHENDE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3020 | GenBank | WP_032490308 |
| Name | WP_032490308_pB-4515_1 |
UniProt ID | _ |
| Length | 133 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 133 a.a. Molecular weight: 15514.81 Da Isoelectric Point: 10.6555
MSLKRFGTRSATGKMVKLKLPVDVEKLLREASDRSGRSRSFEAVIRLKDHLHRYPKFNRKGNVYGKSLVK
YLTMRLDDETNQLLITAKNRSGWCKTDEAADRVIDHLTKFPDFYNSDIFREANEGKKSTFKER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 6477 | GenBank | WP_249895805 |
| Name | traC_LRM91_RS22875_pB-4515_1 |
UniProt ID | _ |
| Length | 882 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 882 a.a. Molecular weight: 100529.30 Da Isoelectric Point: 6.7509
MKNLLDTVTQAANSLLTALKLPDESAQANQILGEMSFPQFSRLLPFRDYNQESGLFMNDATMGFMLEATP
VNGANETIALSLDHLLRTKLPRGIPLCIHLMSSQLVGERIEYGLREFSWSGEQAERFNAITRAYYMKAAE
THFPLPEGMNLPLTLRHYRVFISYCAPSRKKSRADILEMENLVKIIRASLQGAYISTQTVDAQAFINIVG
EMINHNPESLHPTRRRLDPYSDLNYQCVDDSFDLKVRQDYLTLGLRDHGRNSTARIMSFHLARNPEIAFL
WNSADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGD
LRQRLASNQSSVVSYFFNITAFCRDNKETALETEQDILNSFRKNGFELISPRFNHMRNFLACLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITNIDQSAERIRDQLSV
MASPNGNLDEVHEGLLLQAVRAAWLTKKNRARIDDVVDFLENARTSEQYAESPGIRSRLDEMIVLLNQYT
AAGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNRKVGDFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFPQLHREMIEKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLQEHETFRRTYEYRAETD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 52558..71554
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LRM91_RS22770 (LRM91_22770) | 47787..48029 | + | 243 | WP_000131519 | DUF905 domain-containing protein | - |
| LRM91_RS22775 (LRM91_22775) | 48098..50095 | + | 1998 | WP_249895801 | ParB/RepB/Spo0J family partition protein | - |
| LRM91_RS22780 (LRM91_22780) | 50135..50269 | + | 135 | Protein_65 | conjugation system SOS inhibitor PsiB family protein | - |
| LRM91_RS22785 (LRM91_22785) | 50263..50919 | + | 657 | Protein_66 | plasmid SOS inhibition protein A | - |
| LRM91_RS23090 | 50954..51082 | + | 129 | WP_000372430 | hypothetical protein | - |
| LRM91_RS22790 (LRM91_22790) | 51168..51326 | + | 159 | WP_001704449 | hypothetical protein | - |
| LRM91_RS22795 (LRM91_22795) | 51429..52247 | + | 819 | Protein_69 | DUF932 domain-containing protein | - |
| LRM91_RS22800 (LRM91_22800) | 52558..53028 | - | 471 | WP_024134052 | transglycosylase SLT domain-containing protein | virB1 |
| LRM91_RS22805 (LRM91_22805) | 53451..53831 | + | 381 | WP_249895802 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LRM91_RS22810 (LRM91_22810) | 54026..54715 | + | 690 | WP_249895803 | conjugal transfer transcriptional regulator TraJ | - |
| LRM91_RS22815 (LRM91_22815) | 54814..55209 | + | 396 | WP_001746564 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| LRM91_RS22820 (LRM91_22820) | 55251..55613 | + | 363 | WP_001274202 | type IV conjugative transfer system pilin TraA | - |
| LRM91_RS22825 (LRM91_22825) | 55628..55939 | + | 312 | WP_000012129 | type IV conjugative transfer system protein TraL | traL |
| LRM91_RS22830 (LRM91_22830) | 55961..56527 | + | 567 | WP_000399760 | type IV conjugative transfer system protein TraE | traE |
| LRM91_RS22835 (LRM91_22835) | 56514..57254 | + | 741 | WP_015059453 | type-F conjugative transfer system secretin TraK | traK |
| LRM91_RS22840 (LRM91_22840) | 57254..58675 | + | 1422 | WP_000146698 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LRM91_RS22845 (LRM91_22845) | 58665..59261 | + | 597 | WP_000527898 | conjugal transfer pilus-stabilizing protein TraP | - |
| LRM91_RS22850 (LRM91_22850) | 59239..59463 | + | 225 | WP_001746950 | DUF2689 domain-containing protein | - |
| LRM91_RS22855 (LRM91_22855) | 59460..59975 | + | 516 | WP_001015676 | type IV conjugative transfer system lipoprotein TraV | traV |
| LRM91_RS22860 (LRM91_22860) | 60110..60331 | + | 222 | WP_001278699 | conjugal transfer protein TraR | - |
| LRM91_RS22865 (LRM91_22865) | 60324..60791 | + | 468 | WP_249895804 | hypothetical protein | - |
| LRM91_RS22870 (LRM91_22870) | 60788..61171 | + | 384 | WP_000870325 | hypothetical protein | - |
| LRM91_RS22875 (LRM91_22875) | 61187..63835 | + | 2649 | WP_249895805 | type IV secretion system protein TraC | virb4 |
| LRM91_RS22880 (LRM91_22880) | 63810..64196 | + | 387 | WP_000102756 | type-F conjugative transfer system protein TrbI | - |
| LRM91_RS22885 (LRM91_22885) | 64193..64825 | + | 633 | WP_249895806 | type-F conjugative transfer system protein TraW | traW |
| LRM91_RS22890 (LRM91_22890) | 64822..65814 | + | 993 | WP_249895807 | conjugal transfer pilus assembly protein TraU | traU |
| LRM91_RS22895 (LRM91_22895) | 65833..66141 | + | 309 | WP_001576663 | hypothetical protein | - |
| LRM91_RS22900 (LRM91_22900) | 66126..66764 | + | 639 | WP_001576661 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LRM91_RS22905 (LRM91_22905) | 66801..67220 | + | 420 | WP_001229398 | HNH endonuclease | - |
| LRM91_RS22910 (LRM91_22910) | 67217..69043 | + | 1827 | WP_249895808 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LRM91_RS22915 (LRM91_22915) | 69054..69530 | + | 477 | WP_001746543 | hypothetical protein | - |
| LRM91_RS22920 (LRM91_22920) | 69541..69756 | + | 216 | WP_001746544 | conjugal transfer protein TrbE | - |
| LRM91_RS22925 (LRM91_22925) | 69769..70512 | + | 744 | Protein_95 | type-F conjugative transfer system pilin assembly protein TraF | - |
| LRM91_RS22930 (LRM91_22930) | 70726..71019 | + | 294 | WP_001233883 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LRM91_RS22935 (LRM91_22935) | 71006..71554 | + | 549 | WP_249895809 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LRM91_RS22940 (LRM91_22940) | 71544..72815 | + | 1272 | Protein_98 | conjugal transfer pilus assembly protein TraH | - |
| LRM91_RS22945 (LRM91_22945) | 74052..74534 | + | 483 | Protein_99 | surface exclusion protein | - |
| LRM91_RS22950 (LRM91_22950) | 74780..75511 | + | 732 | WP_249895811 | conjugal transfer complement resistance protein TraT | - |
Host bacterium
| ID | 9372 | GenBank | NZ_CP088137 |
| Plasmid name | pB-4515_1 | Incompatibility group | IncFII |
| Plasmid size | 86811 bp | Coordinate of oriT [Strand] | 53102..53163 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Typhimurium strain SCPM-O-B-4515 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |