Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108836 |
| Name | oriT_p2075-1 |
| Organism | Citrobacter freundii strain 2075 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP119166 (41593..41691 [-], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 77..82, 89..94 (AAAAAA..TTTTTT) 77..82, 88..93 (AAAAAA..TTTTTT) 31..38, 41..48 (AGCGTGAT..ATCACGCT) 17..23, 35..41 (TAAATCA..TGATTTA) |
| Location of nic site | 59..60 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
>oriT_p2075-1
TTTGTTTTTTTTCTTTTAAATCAGTGCGATAGCGTGATTTATCACGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTTGGTAG
TTTGTTTTTTTTCTTTTAAATCAGTGCGATAGCGTGATTTATCACGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file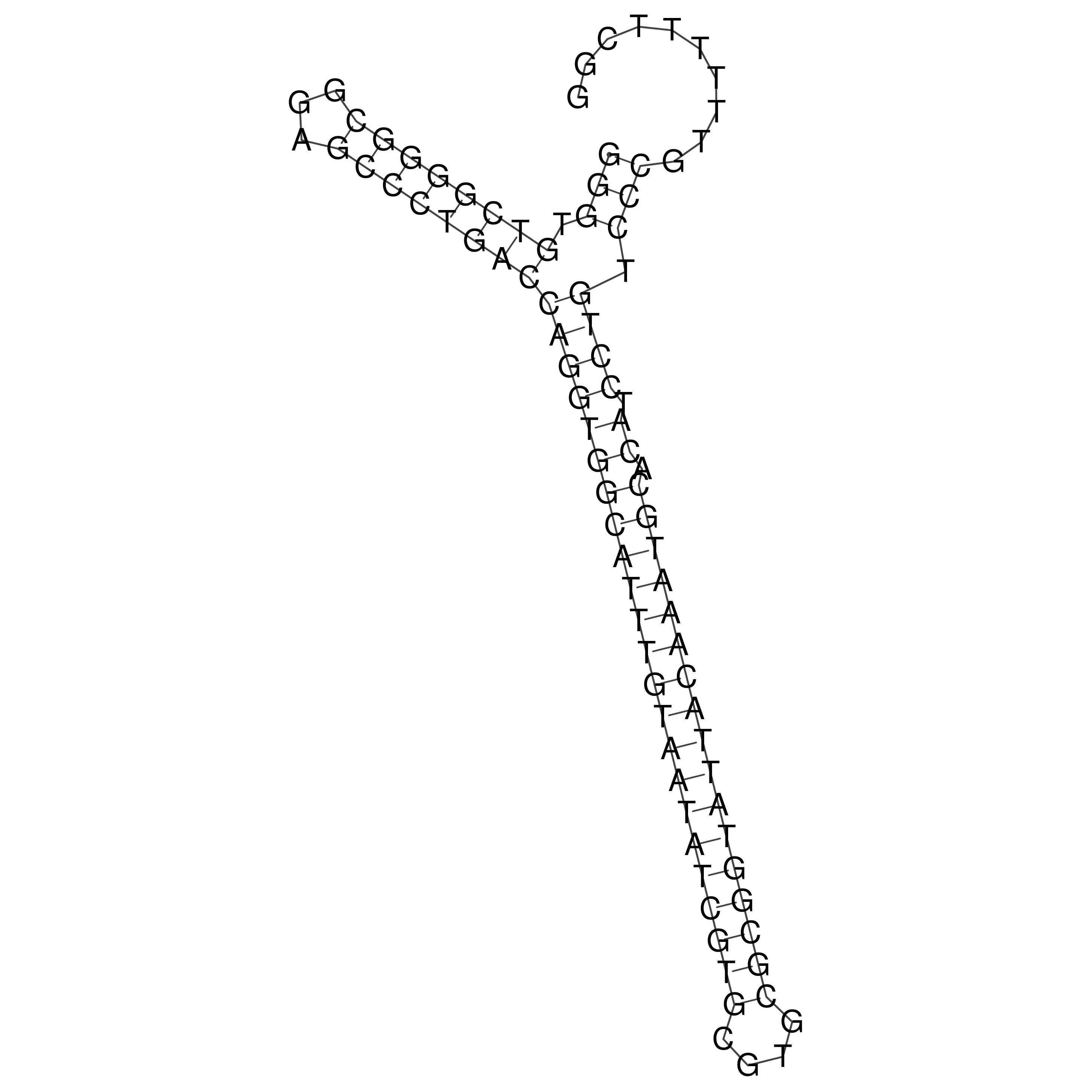
Relaxase
| ID | 5923 | GenBank | WP_000130000 |
| Name | Replic_Relax_P0S03_RS24505_p2075-1 |
UniProt ID | R4WML4 |
| Length | 101 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 101 a.a. Molecular weight: 11477.11 Da Isoelectric Point: 7.5204
>WP_000130000.1 MULTISPECIES: PadR family transcriptional regulator [Pseudomonadota]
MTDKDLYGGLIRLHILHHAAEEPVFGLGIIEELRRHGYEMSAGTVYPMLHGLEKKGYLTSRHERTGRRER
RVYDITEQGRTALADAKTKVKELFGELVEGG
MTDKDLYGGLIRLHILHHAAEEPVFGLGIIEELRRHGYEMSAGTVYPMLHGLEKKGYLTSRHERTGRRER
RVYDITEQGRTALADAKTKVKELFGELVEGG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | R4WML4 |
Host bacterium
| ID | 9271 | GenBank | NZ_CP119166 |
| Plasmid name | p2075-1 | Incompatibility group | IncR |
| Plasmid size | 46049 bp | Coordinate of oriT [Strand] | 41593..41691 [-] |
| Host baterium | Citrobacter freundii strain 2075 |
Cargo genes
| Drug resistance gene | blaKPC-2, blaNDM-1 |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merC, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |