Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108824 |
| Name | oriT_PS_Ent_2_2022|p2 |
| Organism | Salmonella enterica subsp. enterica serovar Enteritidis strain PS_Ent_2_2022 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP120320 (33285..33891 [+], 607 nt) |
| oriT length | 607 nt |
| IRs (inverted repeats) | 590..595, 600..605 (ATGCCC..GGGCAT) 517..523, 529..535 (CCTCCCG..CGGGAGG) 453..458, 462..467 (GTTCGC..GCGAAC) 196..202, 204..210 (TCAATTC..GAATTGA) 175..180, 187..192 (AAATCA..TGATTT) 103..108, 120..125 (GCTAAA..TTTAGC) 79..84, 93..98 (GCTTTA..TAAAGC) 29..37, 46..54 (TTTTGATAA..TTATCAAAA) |
| Location of nic site | 313..314 |
| Conserved sequence flanking the nic site |
TCCTGCATCG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 607 nt
>oriT_PS_Ent_2_2022|p2
CTTTGTTTACCTGTTAAGTACATCGTTGTTTTGATAAAATCATTATTATCAAAAAACGTATTTATATCCTTACTGACAGCTTTACTTTTTAATAAAGCATTTGCTAAAAGTTTCATGCCTTTAGCTACTGTTAAAGCTGGTTCATCCGGATAAAGAGTTTTAAGGCAATCCAGCAAATCATCATGCTGATTTTCATCAATTCTGAATTGAACTTTTTTCATAAAATTCTCCAACAAGAACCGACTGTAGGTCACCGGGCAAATGTTGCGGAATGGCGTCAGAGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAATCTGGCGCCGCTCATAAATTGTGCTTGAGCATCAACAAAAATGCAAAAAGGCTGATGTTGTCATGGCTGAACATACGTACGTCTGCGACCGGAAACCCGATCAATGCGGTAAGGACAGTGCCGTCAGGTTCGCTCCGCGAACGGTCTGGCAGGAAGCGCAAGCGCAGTCCCGCGCAATATATCAGAAAGCACCTCCCGAAATACGGGAGGGCTTCGGCTATTCAGCTAAGGTTGCTCTGTTCAGGTTGCTGAGTGTAGCCTTCCATGCCCTGACGGGCATCA
CTTTGTTTACCTGTTAAGTACATCGTTGTTTTGATAAAATCATTATTATCAAAAAACGTATTTATATCCTTACTGACAGCTTTACTTTTTAATAAAGCATTTGCTAAAAGTTTCATGCCTTTAGCTACTGTTAAAGCTGGTTCATCCGGATAAAGAGTTTTAAGGCAATCCAGCAAATCATCATGCTGATTTTCATCAATTCTGAATTGAACTTTTTTCATAAAATTCTCCAACAAGAACCGACTGTAGGTCACCGGGCAAATGTTGCGGAATGGCGTCAGAGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAATCTGGCGCCGCTCATAAATTGTGCTTGAGCATCAACAAAAATGCAAAAAGGCTGATGTTGTCATGGCTGAACATACGTACGTCTGCGACCGGAAACCCGATCAATGCGGTAAGGACAGTGCCGTCAGGTTCGCTCCGCGAACGGTCTGGCAGGAAGCGCAAGCGCAGTCCCGCGCAATATATCAGAAAGCACCTCCCGAAATACGGGAGGGCTTCGGCTATTCAGCTAAGGTTGCTCTGTTCAGGTTGCTGAGTGTAGCCTTCCATGCCCTGACGGGCATCA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file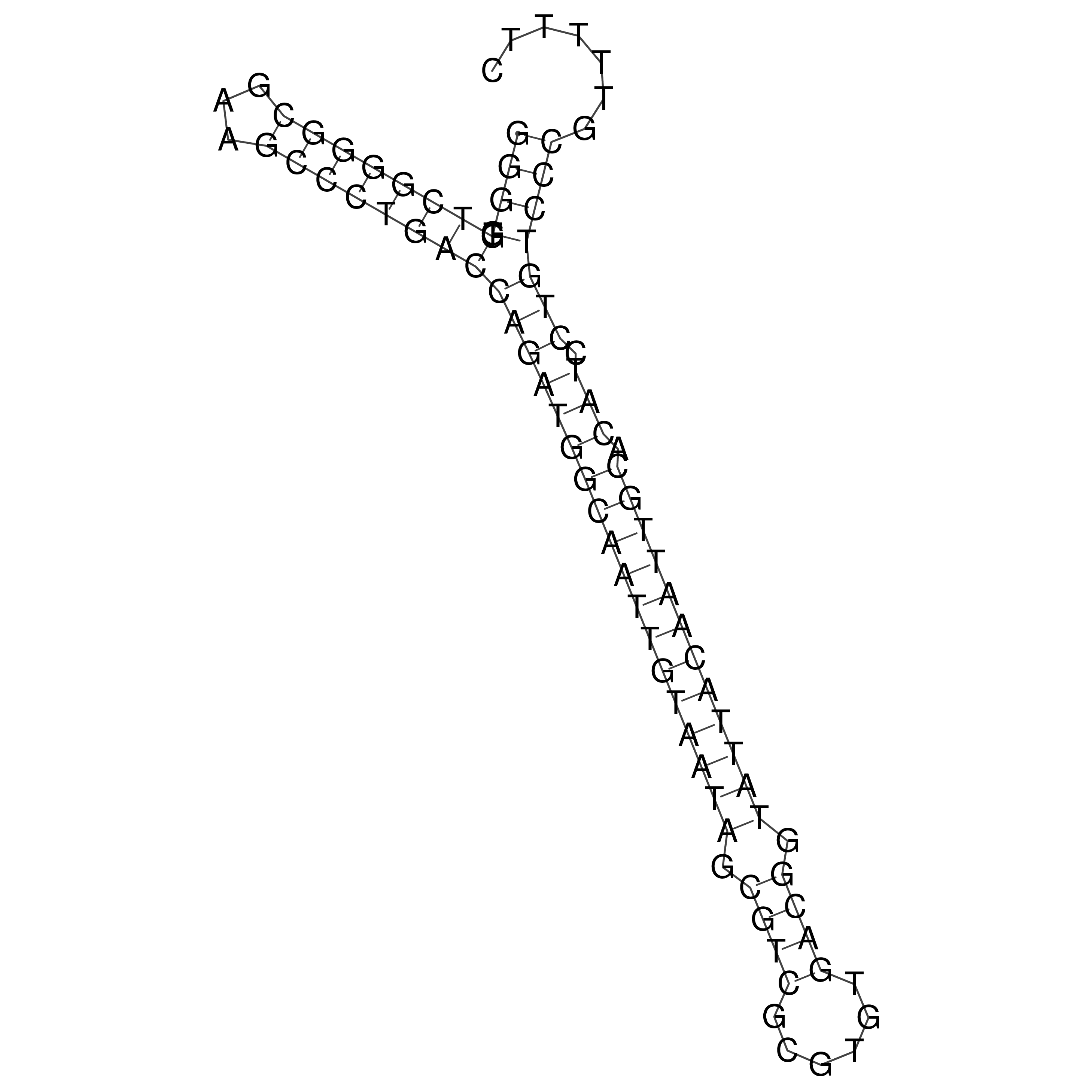
Relaxase
| ID | 5911 | GenBank | WP_000539532 |
| Name | mobP1_P1703_RS23580_PS_Ent_2_2022|p2 |
UniProt ID | K4NPN4 |
| Length | 388 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 388 a.a. Molecular weight: 44348.73 Da Isoelectric Point: 10.5588
>WP_000539532.1 MULTISPECIES: MobP1 family relaxase [Gammaproteobacteria]
MGVYVDKEHRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAVTRQGIRNSI
DYMSRESELPVMSESGRVWTGDEILEAKEHMIDRANDPQHVMNDKGKENKKITQNIVFSPPVSAKVKPED
LLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLKGYD
VKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDIGYDHYQNDKTKSKQHFIKLKTLNKGVEKTYWGADF
GGLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGVDRTPSASKELVLN
SPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFLGL
MGVYVDKEHRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAVTRQGIRNSI
DYMSRESELPVMSESGRVWTGDEILEAKEHMIDRANDPQHVMNDKGKENKKITQNIVFSPPVSAKVKPED
LLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLKGYD
VKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDIGYDHYQNDKTKSKQHFIKLKTLNKGVEKTYWGADF
GGLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGVDRTPSASKELVLN
SPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFLGL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | K4NPN4 |
T4CP
| ID | 6363 | GenBank | WP_000053816 |
| Name | t4cp2_P1703_RS23510_PS_Ent_2_2022|p2 |
UniProt ID | _ |
| Length | 611 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 611 a.a. Molecular weight: 69371.31 Da Isoelectric Point: 8.0636
>WP_000053816.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Gammaproteobacteria]
MSLKLPDKGQWAFIGLVMCLVTYYTGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFIIWQLNKTAVALYGDAKFASDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVFSI
GSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAKIS
EKLGYITTTSKSTSKNRGRSTSQGESESEARRALVLPQELGTLDFKEEFIILKGENPVKAEKALYYLDPY
FMDRLMKVSPKLASLTMKLNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
MSLKLPDKGQWAFIGLVMCLVTYYTGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFIIWQLNKTAVALYGDAKFASDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVFSI
GSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAKIS
EKLGYITTTSKSTSKNRGRSTSQGESESEARRALVLPQELGTLDFKEEFIILKGENPVKAEKALYYLDPY
FMDRLMKVSPKLASLTMKLNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 20793..30745
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| P1703_RS23490 (P1703_23490) | 15803..16009 | - | 207 | WP_000850859 | hemolysin expression modulator Hha | - |
| P1703_RS23495 (P1703_23495) | 16006..18159 | - | 2154 | WP_065622024 | type IA DNA topoisomerase | - |
| P1703_RS23500 (P1703_23500) | 18246..18548 | - | 303 | WP_024242858 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| P1703_RS23505 (P1703_23505) | 18545..18958 | - | 414 | WP_000722610 | cag pathogenicity island Cag12 family protein | - |
| P1703_RS23510 (P1703_23510) | 18955..20790 | - | 1836 | WP_000053816 | type IV secretory system conjugative DNA transfer family protein | - |
| P1703_RS23515 (P1703_23515) | 20793..21824 | - | 1032 | WP_001058474 | P-type DNA transfer ATPase VirB11 | virB11 |
| P1703_RS23520 (P1703_23520) | 21826..23031 | - | 1206 | WP_065622023 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| P1703_RS23525 (P1703_23525) | 23028..23957 | - | 930 | WP_000783379 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| P1703_RS23530 (P1703_23530) | 23962..24675 | - | 714 | WP_000394613 | type IV secretion system protein | virB8 |
| P1703_RS23540 (P1703_23540) | 24920..25981 | - | 1062 | WP_000796671 | type IV secretion system protein | virB6 |
| P1703_RS23545 (P1703_23545) | 25993..26277 | - | 285 | WP_000748128 | EexN family lipoprotein | - |
| P1703_RS23550 (P1703_23550) | 26292..27041 | - | 750 | WP_000744202 | type IV secretion system protein | - |
| P1703_RS23555 (P1703_23555) | 27052..29805 | - | 2754 | WP_001328100 | VirB3 family type IV secretion system protein | virb4 |
| P1703_RS23560 (P1703_23560) | 29824..30120 | - | 297 | WP_000916182 | TrbC/VirB2 family protein | virB2 |
| P1703_RS23565 (P1703_23565) | 30098..30745 | - | 648 | WP_000953530 | lytic transglycosylase domain-containing protein | virB1 |
| P1703_RS23570 (P1703_23570) | 30778..30990 | - | 213 | WP_000125172 | hypothetical protein | - |
| P1703_RS23575 (P1703_23575) | 30920..31438 | - | 519 | WP_001446885 | transcription termination/antitermination NusG family protein | - |
| P1703_RS23580 (P1703_23580) | 31791..32957 | - | 1167 | WP_000539532 | MobP1 family relaxase | - |
| P1703_RS23585 (P1703_23585) | 32960..33505 | - | 546 | WP_000757693 | DNA distortion polypeptide 1 | - |
| P1703_RS23590 (P1703_23590) | 33831..34103 | - | 273 | WP_000160399 | hypothetical protein | - |
| P1703_RS23595 (P1703_23595) | 34116..34265 | - | 150 | WP_000003880 | hypothetical protein | - |
| P1703_RS23600 (P1703_23600) | 34335..34514 | - | 180 | WP_000439078 | hypothetical protein | - |
| P1703_RS23605 (P1703_23605) | 34557..34886 | - | 330 | WP_000866646 | hypothetical protein | - |
| P1703_RS23610 (P1703_23610) | 34979..35194 | - | 216 | WP_001180116 | hypothetical protein | - |
| P1703_RS23615 (P1703_23615) | 35184..35429 | - | 246 | WP_000356547 | hypothetical protein | - |
Host bacterium
| ID | 9259 | GenBank | NZ_CP120320 |
| Plasmid name | PS_Ent_2_2022|p2 | Incompatibility group | IncX1 |
| Plasmid size | 37698 bp | Coordinate of oriT [Strand] | 33285..33891 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Enteritidis strain PS_Ent_2_2022 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |