Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108772 |
| Name | oriT_X14|3 |
| Organism | Nitrobacter hamburgensis X14 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_007961 (13..46 [+], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_X14|3
TCCAAGGGCGCAGTTATACGTCGCTGACGCGACG
TCCAAGGGCGCAGTTATACGTCGCTGACGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file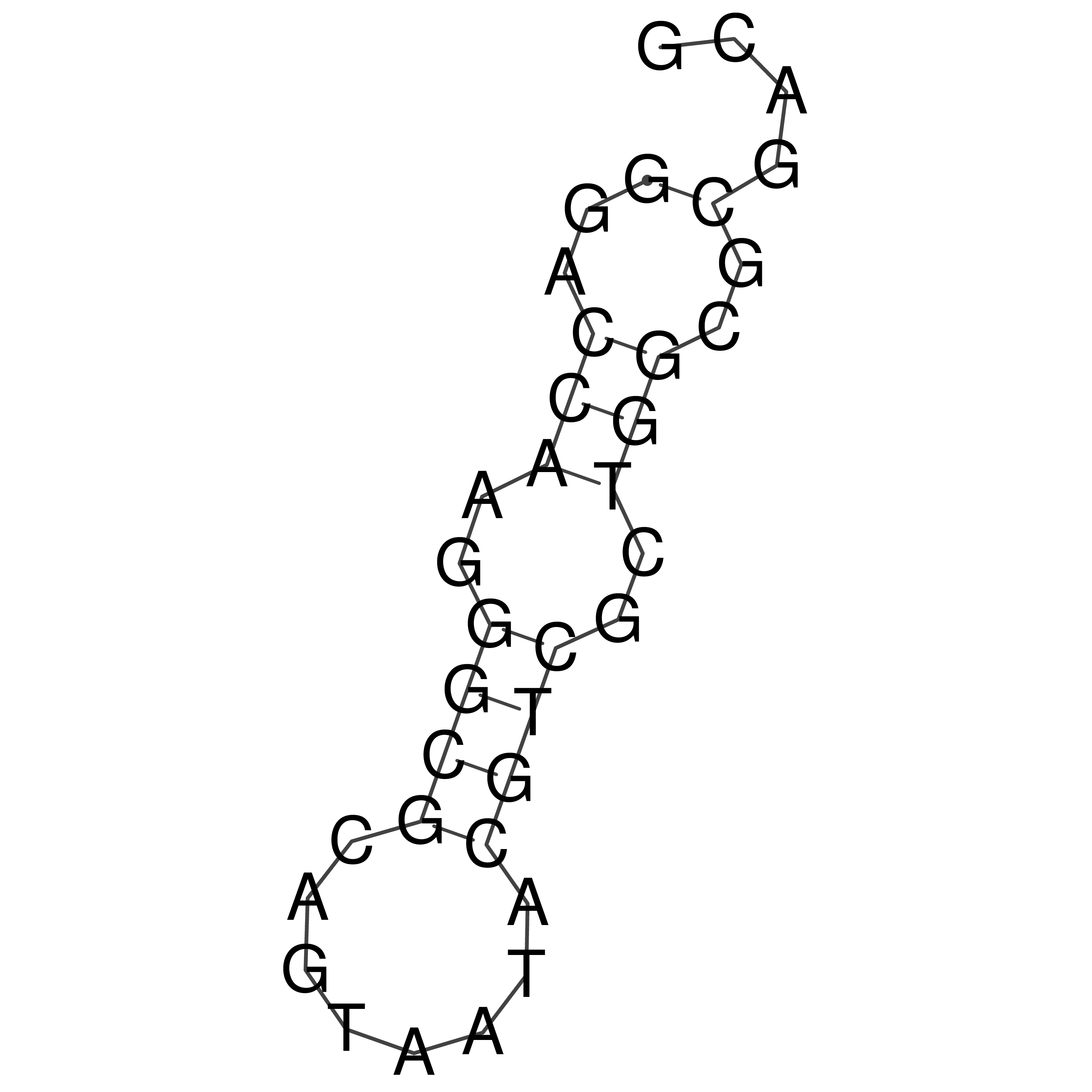
Relaxase
| ID | 5882 | GenBank | WP_011505201 |
| Name | traA_NHAM_RS23140_X14|3 |
UniProt ID | Q1QF79 |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 123275.44 Da Isoelectric Point: 10.3283
>WP_011505201.1 Ti-type conjugative transfer relaxase TraA [Nitrobacter hamburgensis]
MAITHFTVQIVSRGTGGSAVLSAAYRHCARMDYEAEARVVDYSNKRNLAHEDFVLPPDAPAWVRAMIADR
SVSGASETFWNYVEAFETRSDAQFAREAIIALPVELTREQNIALMRAFVTQEILPRGQVADWVYHNEPGN
PHVHLMTTLRPLIESGFGGKKVAVTGEDGKPLRTQAGKIRYALWAGDTPDFLAQRARWIDLQNQHLALAG
LDVRVEGRSYAERGIAIVPTTHIGVATKAIDRRKAKRQGGTQRLDRLAVFEENRAENARRIQRRPEIVLE
LVSREKSVFDERDIAKVLHRYVDDSGTFQNLLTRILQLPEVIRLQAESIDFATGGRAPARYTVRELIRCE
AEMARRAIHLSGSTTFPVKASVIAGVSARHANLSAEQRTAIEHLSSGERIAAVVGRAGAGKTTMMRAARE
MWEEAGYRVVGGALAGKAAEGLEKEAGIAARTLASWELGWKNGRDDLDDKTVFVLDEAGMVASRQMALVV
ETASKAGAKIVLVGDPEQLQPIEAGAAFRSIVERIGYAELETIYRQREQWMRDASLDLARGRTAQALSAY
RQHGRVLGSELKAQAIDSLIADWNRDYDPSKSTLILAHLRRDVRALNDMARAKLVERGLVEEGHVFGTED
GERRFAVGDQIVFLKNEGSLGVKNGMIGKVVDAAPGRLVAEIGDGQGSVENRRRVAVEQRFYRNVDHGYA
TTVHKAQGATVDRVKVLATLSLDKHLTYVALTRHREEVTLYYGRRSFQFAGGLIPILSQRRAKETTLDYE
RGALYRDALRFAANRGLHIVRVARTLINDRRRWTLRQKQRLVDAGRRLRALGERLGLVDSHKPVISTARK
EAKPMVAGVTTFLKSFTDTIEDRILADPAIKKQWKEVSTRFRLVFADPERAFRAVNFDAMLKDSAAAVST
IQQLAANPESFGALRGKTGLLAGKADKQERQRAEANVPALKSDIERYLRLRAEAEHKHEAEERAIRHSVA
IDIPALSRDAHRVLERVRDAIDRNDLPSALEFALADRMVKAEIDGFNKAIAERFGDRTFLPNAAREAKGQ
AFEKAAAGMDPARREQLKEAWPAMRTAQRLAAGERTAQALKQVESQRMMQRQGQTLK
MAITHFTVQIVSRGTGGSAVLSAAYRHCARMDYEAEARVVDYSNKRNLAHEDFVLPPDAPAWVRAMIADR
SVSGASETFWNYVEAFETRSDAQFAREAIIALPVELTREQNIALMRAFVTQEILPRGQVADWVYHNEPGN
PHVHLMTTLRPLIESGFGGKKVAVTGEDGKPLRTQAGKIRYALWAGDTPDFLAQRARWIDLQNQHLALAG
LDVRVEGRSYAERGIAIVPTTHIGVATKAIDRRKAKRQGGTQRLDRLAVFEENRAENARRIQRRPEIVLE
LVSREKSVFDERDIAKVLHRYVDDSGTFQNLLTRILQLPEVIRLQAESIDFATGGRAPARYTVRELIRCE
AEMARRAIHLSGSTTFPVKASVIAGVSARHANLSAEQRTAIEHLSSGERIAAVVGRAGAGKTTMMRAARE
MWEEAGYRVVGGALAGKAAEGLEKEAGIAARTLASWELGWKNGRDDLDDKTVFVLDEAGMVASRQMALVV
ETASKAGAKIVLVGDPEQLQPIEAGAAFRSIVERIGYAELETIYRQREQWMRDASLDLARGRTAQALSAY
RQHGRVLGSELKAQAIDSLIADWNRDYDPSKSTLILAHLRRDVRALNDMARAKLVERGLVEEGHVFGTED
GERRFAVGDQIVFLKNEGSLGVKNGMIGKVVDAAPGRLVAEIGDGQGSVENRRRVAVEQRFYRNVDHGYA
TTVHKAQGATVDRVKVLATLSLDKHLTYVALTRHREEVTLYYGRRSFQFAGGLIPILSQRRAKETTLDYE
RGALYRDALRFAANRGLHIVRVARTLINDRRRWTLRQKQRLVDAGRRLRALGERLGLVDSHKPVISTARK
EAKPMVAGVTTFLKSFTDTIEDRILADPAIKKQWKEVSTRFRLVFADPERAFRAVNFDAMLKDSAAAVST
IQQLAANPESFGALRGKTGLLAGKADKQERQRAEANVPALKSDIERYLRLRAEAEHKHEAEERAIRHSVA
IDIPALSRDAHRVLERVRDAIDRNDLPSALEFALADRMVKAEIDGFNKAIAERFGDRTFLPNAAREAKGQ
AFEKAAAGMDPARREQLKEAWPAMRTAQRLAAGERTAQALKQVESQRMMQRQGQTLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q1QF79 |
T4CP
| ID | 6321 | GenBank | WP_011505221 |
| Name | t4cp2_NHAM_RS23270_X14|3 |
UniProt ID | _ |
| Length | 817 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 817 a.a. Molecular weight: 91398.84 Da Isoelectric Point: 5.4984
>WP_011505221.1 conjugal transfer protein TrbE [Nitrobacter hamburgensis]
MVALRSFRHPSPSFADLVPHAGLVANGIVLLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINTILSRLG
SGWMIQVEAVRVPTTDYPARQDCHFPDPVTRAIDDERRARFEAERGHFESRHAIILTWRPPERRRAGLAR
YIYSDVESRSASYADTAIETFQNAIREIEQYLASVLSIQRMETREVDERDGTRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLATAELQHGLSPMVENRFLGVVAIDGFPAESWPGILNSLDLMPLTYRWSSRFMF
LDDQDARQKLERTRKKWQQKVRPFIDQLFQTQSRSIDQDAAAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDDNSTRLQEKTEAVRRLIQAEGFGARIETLNATEAFLGSLPGNWYANIREPLINTRNLADLIPLNAVWS
GSPTAPCPFYPPNSPPLMQVASGSTPFRMNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYAGAQIFA
FDKGRSMMPLTLAAGGDHYEIGGDDADGSAALAFCPLSDLSTDTDRAWASEWIETLVAVQGVTITPDHRN
AISRQIGLMAEARGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAESDGLELGSFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTRKFYERIGFNDRQIEIVATAVPKREYYVASPEGRRLFNMALGP
VALSFVGASGKDDLKRILALKKDSTDWPVYWLKERGIGHAETLLADS
MVALRSFRHPSPSFADLVPHAGLVANGIVLLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINTILSRLG
SGWMIQVEAVRVPTTDYPARQDCHFPDPVTRAIDDERRARFEAERGHFESRHAIILTWRPPERRRAGLAR
YIYSDVESRSASYADTAIETFQNAIREIEQYLASVLSIQRMETREVDERDGTRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLATAELQHGLSPMVENRFLGVVAIDGFPAESWPGILNSLDLMPLTYRWSSRFMF
LDDQDARQKLERTRKKWQQKVRPFIDQLFQTQSRSIDQDAAAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDDNSTRLQEKTEAVRRLIQAEGFGARIETLNATEAFLGSLPGNWYANIREPLINTRNLADLIPLNAVWS
GSPTAPCPFYPPNSPPLMQVASGSTPFRMNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYAGAQIFA
FDKGRSMMPLTLAAGGDHYEIGGDDADGSAALAFCPLSDLSTDTDRAWASEWIETLVAVQGVTITPDHRN
AISRQIGLMAEARGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAESDGLELGSFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTRKFYERIGFNDRQIEIVATAVPKREYYVASPEGRRLFNMALGP
VALSFVGASGKDDLKRILALKKDSTDWPVYWLKERGIGHAETLLADS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 6322 | GenBank | WP_011505305 |
| Name | traG_NHAM_RS23775_X14|3 |
UniProt ID | _ |
| Length | 641 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 641 a.a. Molecular weight: 68992.73 Da Isoelectric Point: 9.2301
>WP_011505305.1 Ti-type conjugative transfer system protein TraG [Nitrobacter hamburgensis]
MKARKIALAIAPAILMAVLCLATTGAESWLARFGTTPGGRMLLGRIGLALPYALPAAAAVIFLFAAAGAN
AIRAAGWGVVTGGCLVVVIAASREIARLHSFADAVPAGERLLAYADPSTIGGATIAALVAAFGLRVAMRG
NAAFAAAAPRRIRGRRAIHGESDWMAMTEAARLFAEAGGIVVGERYRVDQDAVADRAFRADEADTWGAGG
KSPLLCFDASFGSSHAIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSNEVAPMVLTHRRNAGRQAFVLD
PRSPEGGFNVLDWVGQHGATKEEDIAAVASWIMTDNPRQGSARDDFFRASALQLLTALVADICLSGHTEK
KDQHLRQVRANLSEPEPKLRQRLQAIYDNSASGFVKENVAPFIAMTPETFSGVYANAAKETHWLSYANYA
ALVSGSSFTTAELADGQTDIFINLDLKTLENHPGLARVIIGSLLNAIYNRNGEVSGHTLFLLDEVARLGY
LRILETARDAGRKYGITLLMLYQSIGQMREAYGGRDATSKWFESASWISFSAINDPETADYISKRCGDTT
VEADQLSRSSQMSGSSRTRSKQLARRPLIMPHEVLRMRTDEQIVFTAGNPPLRCGRAIWFRRRDMKACTG
QDPYHRSEAVR
MKARKIALAIAPAILMAVLCLATTGAESWLARFGTTPGGRMLLGRIGLALPYALPAAAAVIFLFAAAGAN
AIRAAGWGVVTGGCLVVVIAASREIARLHSFADAVPAGERLLAYADPSTIGGATIAALVAAFGLRVAMRG
NAAFAAAAPRRIRGRRAIHGESDWMAMTEAARLFAEAGGIVVGERYRVDQDAVADRAFRADEADTWGAGG
KSPLLCFDASFGSSHAIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSNEVAPMVLTHRRNAGRQAFVLD
PRSPEGGFNVLDWVGQHGATKEEDIAAVASWIMTDNPRQGSARDDFFRASALQLLTALVADICLSGHTEK
KDQHLRQVRANLSEPEPKLRQRLQAIYDNSASGFVKENVAPFIAMTPETFSGVYANAAKETHWLSYANYA
ALVSGSSFTTAELADGQTDIFINLDLKTLENHPGLARVIIGSLLNAIYNRNGEVSGHTLFLLDEVARLGY
LRILETARDAGRKYGITLLMLYQSIGQMREAYGGRDATSKWFESASWISFSAINDPETADYISKRCGDTT
VEADQLSRSSQMSGSSRTRSKQLARRPLIMPHEVLRMRTDEQIVFTAGNPPLRCGRAIWFRRRDMKACTG
QDPYHRSEAVR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 15988..25519
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NHAM_RS27830 | 11327..11581 | - | 255 | WP_011504828 | hypothetical protein | - |
| NHAM_RS23215 (Nham_4554) | 11777..11944 | + | 168 | WP_157043862 | hypothetical protein | - |
| NHAM_RS23220 (Nham_4555) | 11949..12368 | + | 420 | WP_011505210 | hypothetical protein | - |
| NHAM_RS23225 (Nham_4556) | 13055..14200 | - | 1146 | WP_011505211 | IS110 family transposase | - |
| NHAM_RS28730 | 14494..14670 | - | 177 | WP_245270143 | hypothetical protein | - |
| NHAM_RS23230 (Nham_4558) | 14844..15548 | - | 705 | WP_011505213 | autoinducer binding domain-containing protein | - |
| NHAM_RS23235 (Nham_4559) | 15988..17289 | - | 1302 | WP_011505214 | IncP-type conjugal transfer protein TrbI | virB10 |
| NHAM_RS23240 (Nham_4560) | 17300..17752 | - | 453 | WP_011505215 | conjugal transfer protein TrbH | - |
| NHAM_RS23245 (Nham_4561) | 17755..18594 | - | 840 | WP_011505216 | P-type conjugative transfer protein TrbG | virB9 |
| NHAM_RS23250 (Nham_4562) | 18612..19274 | - | 663 | WP_011505217 | conjugal transfer protein TrbF | virB8 |
| NHAM_RS23255 (Nham_4563) | 19317..20471 | - | 1155 | WP_011505218 | P-type conjugative transfer protein TrbL | virB6 |
| NHAM_RS27835 (Nham_4564) | 20484..20669 | - | 186 | WP_011505219 | entry exclusion protein TrbK | - |
| NHAM_RS23265 (Nham_4565) | 20666..21463 | - | 798 | WP_011505220 | P-type conjugative transfer protein TrbJ | virB5 |
| NHAM_RS23270 (Nham_4566) | 21435..23888 | - | 2454 | WP_011505221 | conjugal transfer protein TrbE | virb4 |
| NHAM_RS23275 (Nham_4567) | 23898..24197 | - | 300 | WP_011505222 | conjugal transfer protein TrbD | virB3 |
| NHAM_RS23280 (Nham_4568) | 24190..24546 | - | 357 | WP_086008386 | TrbC/VirB2 family protein | virB2 |
| NHAM_RS23285 (Nham_4569) | 24551..25519 | - | 969 | WP_041359818 | P-type conjugative transfer ATPase TrbB | virB11 |
| NHAM_RS23290 (Nham_4570) | 25537..26160 | - | 624 | WP_011505225 | acyl-homoserine-lactone synthase | - |
| NHAM_RS23295 (Nham_4571) | 26576..27790 | + | 1215 | WP_011505226 | plasmid partitioning protein RepA | - |
| NHAM_RS23300 (Nham_4572) | 27787..28818 | + | 1032 | WP_011505227 | plasmid partitioning protein RepB | - |
| NHAM_RS23305 (Nham_4573) | 29095..30420 | + | 1326 | WP_011505228 | plasmid replication protein RepC | - |
Host bacterium
| ID | 9207 | GenBank | NC_007961 |
| Plasmid name | X14|3 | Incompatibility group | - |
| Plasmid size | 121408 bp | Coordinate of oriT [Strand] | 13..46 [+] |
| Host baterium | Nitrobacter hamburgensis X14 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | RPYSC3_16500 |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF11 |