Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108769 |
| Name | oriT_pDR208 |
| Organism | Pseudomonas viridiflava strain JACO-C-5 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP097287 (17796..18148 [+], 353 nt) |
| oriT length | 353 nt |
| IRs (inverted repeats) | 103..109, 113..119 (GCCGCGC..GCGCGGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 353 nt
>oriT_pDR208
GTATAGCAAATGTAATACACAAAAAGCTTGATTTAGCTGGGCTAGATGGTAGTTTTGTATAGCAAGTGTATAGCGCAAGAAACTACACGCATTTCGCTGTAGGCCGCGCTCTGCGCGGCTTGCGCCTCCCGCTAGGGGGGTGCGTGATGTGTATTGAGATTTTGACCCTGTTAAGGGGAAAAATCGAAGCTCTTGTTAAGGGCGTATCCACTGTTTAGGCCGAAGGCCGTAGGGTGGTATGCAGAAAGGGTGCAGGAAGGGTGCTATAAGAGTGCAGGAGCAGTGCAACTTGACTACAGGTAGGGGGCAGGTAAGATGCACATAGAGTGCAACAGGGGTGCAGGTAGGGTGCA
GTATAGCAAATGTAATACACAAAAAGCTTGATTTAGCTGGGCTAGATGGTAGTTTTGTATAGCAAGTGTATAGCGCAAGAAACTACACGCATTTCGCTGTAGGCCGCGCTCTGCGCGGCTTGCGCCTCCCGCTAGGGGGGTGCGTGATGTGTATTGAGATTTTGACCCTGTTAAGGGGAAAAATCGAAGCTCTTGTTAAGGGCGTATCCACTGTTTAGGCCGAAGGCCGTAGGGTGGTATGCAGAAAGGGTGCAGGAAGGGTGCTATAAGAGTGCAGGAGCAGTGCAACTTGACTACAGGTAGGGGGCAGGTAAGATGCACATAGAGTGCAACAGGGGTGCAGGTAGGGTGCA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file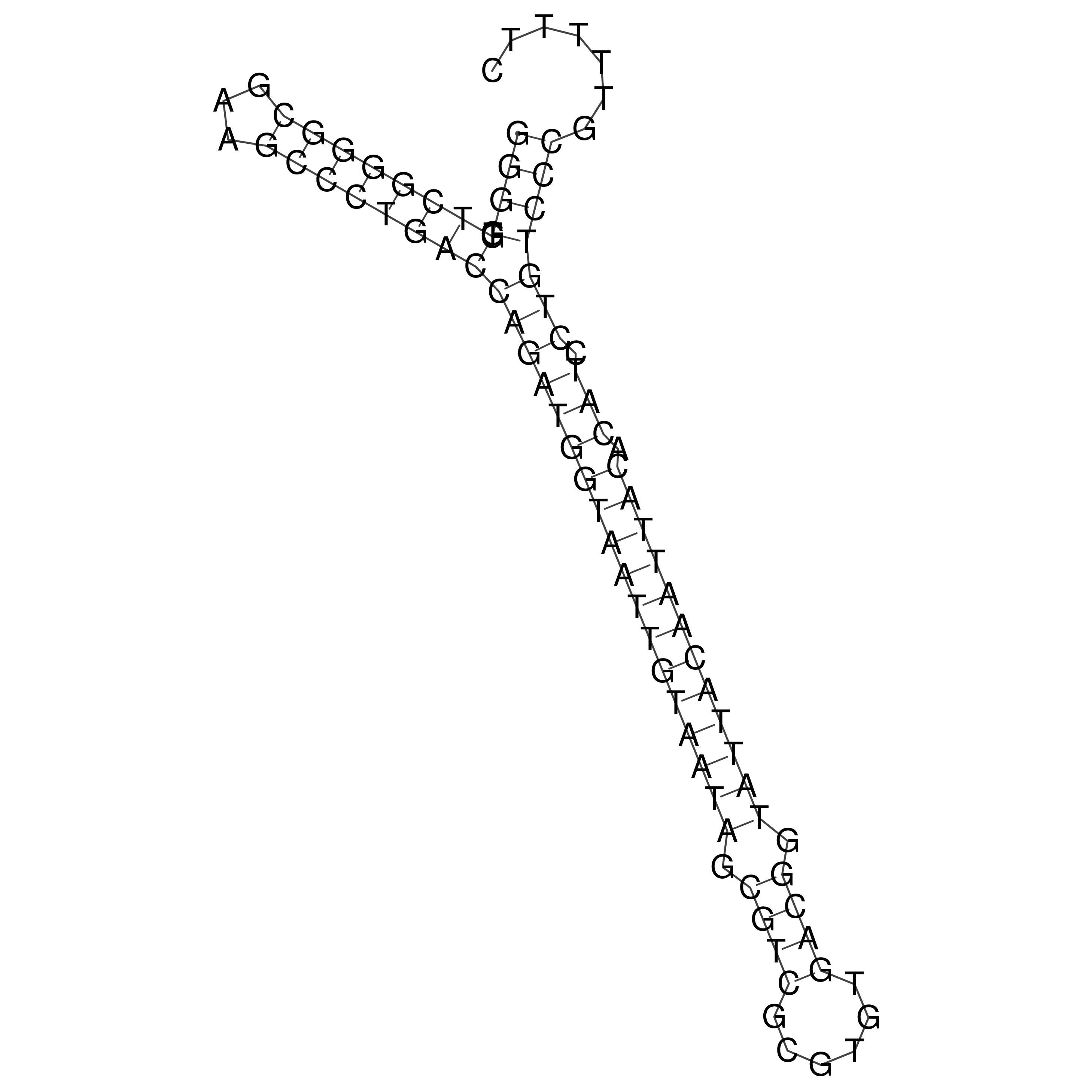
Relaxase
| ID | 5879 | GenBank | WP_054076663 |
| Name | mobF_M4X66_RS27060_pDR208 |
UniProt ID | _ |
| Length | 981 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 981 a.a. Molecular weight: 109217.07 Da Isoelectric Point: 9.9333
>WP_054076663.1 MULTISPECIES: MobF family relaxase [Pseudomonas]
MFNVTSIKGSNQYAAAGYYTAADDYYAKESPGEWQGIGAELLGLEGPIDQQELAKLFDGKLPNGEMMPKP
VNKETGEIVTRRMGLDLTFSAPKSVSMQALIAGDKDVVAAHDRAVTKAMEHVEKLAQTRRKEHGKSMLER
TGNLIIGKFRHELSRAKDPQLHTHAVVMNATRRADGKWRAIHNDDIFKIQPQIDAMYKGELAKELRELGY
EIRVLDKDGNFELAHISRDQIEAFSSRAKVIEDALAKDGKTRSNATALEKQIIAMATRPRKDERDRHLVK
EYWVTKARELGIEFGGRSQLDNREYGRRSESIHAEHNLPEGISAGQAVVQYAINHLTEREQVVGESDLRT
AALRRAVGLASPTEVDDEIKRLVKQGSLIESPPAYRMATAPDGAALSPAGWRALLKEQKGWSEKEAQQYV
KKAIDRGSLVEVEKRYTTQRALKREKAILAIERTGRGQVTPLLTKEQVAKALEGSTLSAGQYQAVEVIVS
TSNRFVGIQGDAGTGKTYSVDRAVKLIESVNNAMTERSTTTDATFRVVALAPYGNQVTALKNEGLDAHTL
ASFFHTKDKKLDERTIVVLDEAGVVGARQMEQLMRIIEQSGARLVQLGDTKQTEAIEAGKPFAQLQQNGM
ETARIKEIQRQKNQELKIAVQHAAEGNPGKSLAHVNHVEEFRDPALRHEAIVRDYVSLTPEERKEVLIVA
GTNKDRKQINTMTREALGLVGKGLELPTLNRVDTTQAERRYAPSYKKGMIVQPEKDYIRAGLSRGELYTV
DQALPGNVLVVKDKNGNRVEFNPRKLTKLSVYNLEKPEFSVGDLVRITRNDQKLDLTNGDRMRVVGNVNG
VIELASVKERDGKPERVVTLPTNRPLHLEHAYSATVHSAQGLTNDRVMISINTKSRTTSQNLWYVAISRA
RYEARIYTDKIAHLPAAIANRYDKTTALSLQQERERQRKEPIKPRTVLDGKAMEKKQRTALDGASAGKSG
V
MFNVTSIKGSNQYAAAGYYTAADDYYAKESPGEWQGIGAELLGLEGPIDQQELAKLFDGKLPNGEMMPKP
VNKETGEIVTRRMGLDLTFSAPKSVSMQALIAGDKDVVAAHDRAVTKAMEHVEKLAQTRRKEHGKSMLER
TGNLIIGKFRHELSRAKDPQLHTHAVVMNATRRADGKWRAIHNDDIFKIQPQIDAMYKGELAKELRELGY
EIRVLDKDGNFELAHISRDQIEAFSSRAKVIEDALAKDGKTRSNATALEKQIIAMATRPRKDERDRHLVK
EYWVTKARELGIEFGGRSQLDNREYGRRSESIHAEHNLPEGISAGQAVVQYAINHLTEREQVVGESDLRT
AALRRAVGLASPTEVDDEIKRLVKQGSLIESPPAYRMATAPDGAALSPAGWRALLKEQKGWSEKEAQQYV
KKAIDRGSLVEVEKRYTTQRALKREKAILAIERTGRGQVTPLLTKEQVAKALEGSTLSAGQYQAVEVIVS
TSNRFVGIQGDAGTGKTYSVDRAVKLIESVNNAMTERSTTTDATFRVVALAPYGNQVTALKNEGLDAHTL
ASFFHTKDKKLDERTIVVLDEAGVVGARQMEQLMRIIEQSGARLVQLGDTKQTEAIEAGKPFAQLQQNGM
ETARIKEIQRQKNQELKIAVQHAAEGNPGKSLAHVNHVEEFRDPALRHEAIVRDYVSLTPEERKEVLIVA
GTNKDRKQINTMTREALGLVGKGLELPTLNRVDTTQAERRYAPSYKKGMIVQPEKDYIRAGLSRGELYTV
DQALPGNVLVVKDKNGNRVEFNPRKLTKLSVYNLEKPEFSVGDLVRITRNDQKLDLTNGDRMRVVGNVNG
VIELASVKERDGKPERVVTLPTNRPLHLEHAYSATVHSAQGLTNDRVMISINTKSRTTSQNLWYVAISRA
RYEARIYTDKIAHLPAAIANRYDKTTALSLQQERERQRKEPIKPRTVLDGKAMEKKQRTALDGASAGKSG
V
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 6318 | GenBank | WP_054076662 |
| Name | t4cp2_M4X66_RS27065_pDR208 |
UniProt ID | _ |
| Length | 518 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 518 a.a. Molecular weight: 58181.56 Da Isoelectric Point: 8.9119
>WP_054076662.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Pseudomonas]
MHKREVDSATMTAGFGLPIAGWGAGVYLAQMPLTPFPAAFKETLHNGWHNPIVLGATIATSVAAASLVYY
LQEYCDDGFRGEQYQEFLRGSQIKNWHTVKAKVNRRNAQTNRERKKKGLAKSDPIMIGKLPMPLHLEDRN
TMICASIGAGKSVTMEGMIASALKRGDRMAVVDPNGTFYSKFSFKGDYILNPFDVRSAGWTIFNEIRGIH
DFNRMAKSIIPPQVDPSDEQWCAYARDVLSDTMRKLKETNNPNQDTLVNLLVREDGDTIRAFLANTDSEG
YFRDNAEKAIASIQFMMNKYIRPLRYMSKGEFSIYQWVNDPNAGNLFITWREDMRDTMKPLVATWIDTIC
ATILSSEPMTGKRLWLFLDELQSLGKLESFVPAATKGRKHGLRMVGSLQDWSQLNASYGRDDAETLLSCF
RNYVILAAANAKNAGMASEILGSHDVKRWRKSLTAGKLTRTEEVTRDEPVVQASEISNLPDLVAYVKFGE
DFPITKVKTPYIDYKERAQAIMITGQAA
MHKREVDSATMTAGFGLPIAGWGAGVYLAQMPLTPFPAAFKETLHNGWHNPIVLGATIATSVAAASLVYY
LQEYCDDGFRGEQYQEFLRGSQIKNWHTVKAKVNRRNAQTNRERKKKGLAKSDPIMIGKLPMPLHLEDRN
TMICASIGAGKSVTMEGMIASALKRGDRMAVVDPNGTFYSKFSFKGDYILNPFDVRSAGWTIFNEIRGIH
DFNRMAKSIIPPQVDPSDEQWCAYARDVLSDTMRKLKETNNPNQDTLVNLLVREDGDTIRAFLANTDSEG
YFRDNAEKAIASIQFMMNKYIRPLRYMSKGEFSIYQWVNDPNAGNLFITWREDMRDTMKPLVATWIDTIC
ATILSSEPMTGKRLWLFLDELQSLGKLESFVPAATKGRKHGLRMVGSLQDWSQLNASYGRDDAETLLSCF
RNYVILAAANAKNAGMASEILGSHDVKRWRKSLTAGKLTRTEEVTRDEPVVQASEISNLPDLVAYVKFGE
DFPITKVKTPYIDYKERAQAIMITGQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 46..17349
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| M4X66_RS26985 | 46..2688 | + | 2643 | WP_081003828 | type VI secretion protein | virb4 |
| M4X66_RS26990 | 2688..3371 | + | 684 | WP_042934861 | type IV secretion system protein | virB5 |
| M4X66_RS26995 | 3399..3611 | + | 213 | WP_017124976 | hypothetical protein | - |
| M4X66_RS27000 | 3604..4008 | + | 405 | WP_054076671 | hypothetical protein | - |
| M4X66_RS27005 | 3996..4859 | + | 864 | WP_017124978 | type IV secretion system protein | virB6 |
| M4X66_RS27010 | 4859..5524 | + | 666 | WP_054076670 | type IV secretion system protein | virB8 |
| M4X66_RS27015 | 5551..6342 | + | 792 | WP_054076669 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| M4X66_RS27020 | 6345..7628 | + | 1284 | WP_054076668 | TrbI/VirB10 family protein | virB10 |
| M4X66_RS27025 | 7638..8666 | + | 1029 | WP_017124982 | P-type DNA transfer ATPase VirB11 | virB11 |
| M4X66_RS27030 | 8650..9552 | + | 903 | WP_054076667 | lytic transglycosylase domain-containing protein | virB1 |
| M4X66_RS27035 | 9582..9911 | + | 330 | WP_054076666 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| M4X66_RS27040 | 9901..10506 | + | 606 | WP_081003826 | phospholipase D family protein | - |
| M4X66_RS27045 | 10521..11003 | + | 483 | WP_054076665 | hypothetical protein | - |
| M4X66_RS27050 | 11048..11686 | + | 639 | WP_054076664 | hypothetical protein | - |
| M4X66_RS27055 | 12009..12764 | - | 756 | WP_230847846 | hypothetical protein | - |
| M4X66_RS27060 | 12837..15782 | - | 2946 | WP_054076663 | MobF family relaxase | - |
| M4X66_RS27065 | 15793..17349 | - | 1557 | WP_054076662 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| M4X66_RS27070 | 17342..17734 | - | 393 | WP_017124989 | hypothetical protein | - |
| M4X66_RS27075 | 18151..18606 | + | 456 | WP_169880829 | conjugal transfer protein TraD | - |
| M4X66_RS27080 | 18603..19322 | + | 720 | WP_054076660 | StbB family protein | - |
| M4X66_RS27085 | 19326..19712 | + | 387 | WP_054076659 | hypothetical protein | - |
| M4X66_RS27090 | 19845..20648 | + | 804 | WP_054076658 | SprT-like domain-containing protein | - |
| M4X66_RS27095 | 21387..21650 | - | 264 | WP_081003825 | antirestriction protein ArdR | - |
| M4X66_RS27100 | 21905..22177 | - | 273 | WP_054076656 | hypothetical protein | - |
Host bacterium
| ID | 9204 | GenBank | NZ_CP097287 |
| Plasmid name | pDR208 | Incompatibility group | - |
| Plasmid size | 42253 bp | Coordinate of oriT [Strand] | 17796..18148 [+] |
| Host baterium | Pseudomonas viridiflava strain JACO-C-5 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |