Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108567 |
| Name | oriT_pBL708 |
| Organism | Salmonella enterica subsp. enterica serovar 14[5]12:i:- strain BL708 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP116043 (35841..36447 [+], 607 nt) |
| oriT length | 607 nt |
| IRs (inverted repeats) | 590..595, 600..605 (ATGCCC..GGGCAT) 517..523, 529..535 (CCTCCCG..CGGGAGG) 453..458, 462..467 (GTTCGC..GCGAAC) 196..202, 204..210 (TCAATTC..GAATTGA) 175..180, 187..192 (AAATCA..TGATTT) 170..175, 184..189 (TCAGCA..TGCTGA) 103..108, 120..125 (GCTAAA..TTTAGC) 79..84, 93..98 (GCTTTA..TAAAGC) 29..37, 46..54 (TTTTGATAA..TTATCAAAA) |
| Location of nic site | 313..314 |
| Conserved sequence flanking the nic site |
TCCTGCATCG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 607 nt
>oriT_pBL708
CTTTGTTTACCTGTTAAGTACATCGTTGTTTTGATAAAATCATTATTATCAAAAAACGTATTTATATCCTTACTGACAGCTTTACTTTTTAATAAAGCATTTGCTAAAAGTTTCATGCCTTTAGCTACTGTTAAAGCTGGTTCATCCGGATAAAGAGTTTTAAGGCAATTCAGCAAATCATCATGCTGATTTTCATCAATTCTGAATTGAACTTTTTTCATAAAATTCTCCGACAAGAACCGACTGTAGGTCACCGGGCAAATGTTGCGGAATGGCGTCAGAGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAATCTGGCGCCGCTCATAAATTGTGCTTGAGCATCAACAAAAATGCAAAAAGGCTGATGTTGTCATGGCTGAACATACGTGCGTCTGCGACCAGAAACTCGATCAATGCCGTAAGGACAGTGCCGTCAGGTTCGCTCCGCGAACGGTCTGGCAGGAAGCGCAAGCGCAGTCCCGCGCAATATATCAGAAAGCACCTCCCGAAATACGGGAGGGCTTCGGCTATTCAGCTAAGGTTGCTCTGTTCAGGTTGCTGAGTGTAGCCTTCCATGCCCTGACGGGCATCA
CTTTGTTTACCTGTTAAGTACATCGTTGTTTTGATAAAATCATTATTATCAAAAAACGTATTTATATCCTTACTGACAGCTTTACTTTTTAATAAAGCATTTGCTAAAAGTTTCATGCCTTTAGCTACTGTTAAAGCTGGTTCATCCGGATAAAGAGTTTTAAGGCAATTCAGCAAATCATCATGCTGATTTTCATCAATTCTGAATTGAACTTTTTTCATAAAATTCTCCGACAAGAACCGACTGTAGGTCACCGGGCAAATGTTGCGGAATGGCGTCAGAGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAATCTGGCGCCGCTCATAAATTGTGCTTGAGCATCAACAAAAATGCAAAAAGGCTGATGTTGTCATGGCTGAACATACGTGCGTCTGCGACCAGAAACTCGATCAATGCCGTAAGGACAGTGCCGTCAGGTTCGCTCCGCGAACGGTCTGGCAGGAAGCGCAAGCGCAGTCCCGCGCAATATATCAGAAAGCACCTCCCGAAATACGGGAGGGCTTCGGCTATTCAGCTAAGGTTGCTCTGTTCAGGTTGCTGAGTGTAGCCTTCCATGCCCTGACGGGCATCA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file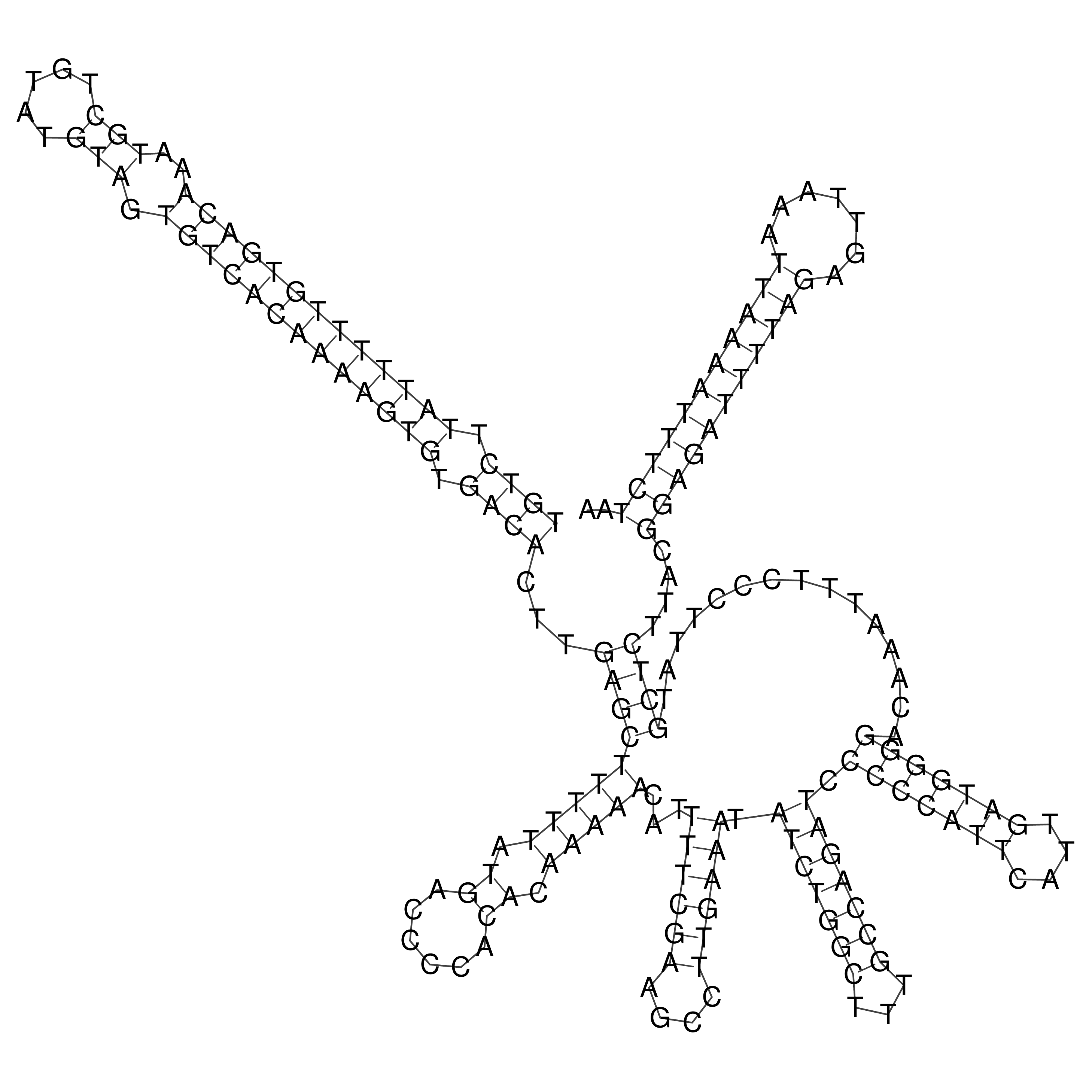
Relaxase
| ID | 5715 | GenBank | WP_000539529 |
| Name | mobP1_PGC28_RS24755_pBL708 |
UniProt ID | A0A0F7JE92 |
| Length | 388 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 388 a.a. Molecular weight: 44493.85 Da Isoelectric Point: 10.5286
>WP_000539529.1 MULTISPECIES: MobP1 family relaxase [Enterobacteriaceae]
MGVYVDKEHRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAITRQGIRNSI
DYMSRESELPVMSESGRVWTGDEILEAKEHMIDRANDPQHVMNDKGKENKKITQNIVFSPPVSAKVKPED
LLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLKGYD
VKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDIGYDHYQNDKTKSKQHFIKLKTLNKGVEKTYWGADF
GDLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGVDRTPSASKELVLN
SPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFWGL
MGVYVDKEHRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAITRQGIRNSI
DYMSRESELPVMSESGRVWTGDEILEAKEHMIDRANDPQHVMNDKGKENKKITQNIVFSPPVSAKVKPED
LLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLKGYD
VKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDIGYDHYQNDKTKSKQHFIKLKTLNKGVEKTYWGADF
GDLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGVDRTPSASKELVLN
SPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFWGL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0F7JE92 |
T4CP
| ID | 6102 | GenBank | WP_000053818 |
| Name | t4cp2_PGC28_RS24685_pBL708 |
UniProt ID | _ |
| Length | 611 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 611 a.a. Molecular weight: 69331.19 Da Isoelectric Point: 7.4339
>WP_000053818.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MSLKLPDKGQWAFIGLVMCLVTYYTGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFIIWQLNKTDVALYGDAKFASDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVFSI
GSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAKIS
EKLGYITTTSKSTSKSRGRSTSQGESESEARRALVLPQELGTLDFKEEFIILKGANPVKAEKALYYLDPY
FMDRLMKVSPKLASLTMELNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
MSLKLPDKGQWAFIGLVMCLVTYYTGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFIIWQLNKTDVALYGDAKFASDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVFSI
GSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAKIS
EKLGYITTTSKSTSKSRGRSTSQGESESEARRALVLPQELGTLDFKEEFIILKGANPVKAEKALYYLDPY
FMDRLMKVSPKLASLTMELNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 23285..33301
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PGC28_RS24670 (PGC28_24655) | 19700..20623 | + | 924 | WP_000902514 | SMEK domain-containing protein | - |
| PGC28_RS24675 (PGC28_24660) | 20738..21040 | - | 303 | WP_000717624 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| PGC28_RS24680 (PGC28_24665) | 21037..21450 | - | 414 | WP_000722603 | cag pathogenicity island Cag12 family protein | - |
| PGC28_RS24685 (PGC28_24670) | 21447..23282 | - | 1836 | WP_000053818 | type IV secretory system conjugative DNA transfer family protein | - |
| PGC28_RS24690 (PGC28_24675) | 23285..24316 | - | 1032 | WP_001058465 | P-type DNA transfer ATPase VirB11 | virB11 |
| PGC28_RS24695 (PGC28_24680) | 24318..25523 | - | 1206 | WP_000139139 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| PGC28_RS24700 (PGC28_24685) | 25520..26449 | - | 930 | WP_000783379 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| PGC28_RS24705 (PGC28_24690) | 26454..27167 | - | 714 | WP_000394613 | type IV secretion system protein | virB8 |
| PGC28_RS24715 (PGC28_24700) | 27412..28542 | - | 1131 | WP_000524912 | type IV secretion system protein | virB6 |
| PGC28_RS24720 (PGC28_24705) | 28551..28838 | - | 288 | WP_000835347 | EexN family lipoprotein | - |
| PGC28_RS24725 (PGC28_24710) | 28839..29597 | - | 759 | WP_000744200 | type IV secretion system protein | - |
| PGC28_RS24730 (PGC28_24715) | 29608..32361 | - | 2754 | WP_016731577 | VirB3 family type IV secretion system protein | virb4 |
| PGC28_RS24735 (PGC28_24720) | 32380..32676 | - | 297 | WP_000916182 | TrbC/VirB2 family protein | virB2 |
| PGC28_RS24740 (PGC28_24725) | 32654..33301 | - | 648 | WP_000953525 | lytic transglycosylase domain-containing protein | virB1 |
| PGC28_RS24745 (PGC28_24730) | 33334..33546 | - | 213 | WP_000125172 | hypothetical protein | - |
| PGC28_RS24750 (PGC28_24735) | 33476..33994 | - | 519 | WP_001446885 | transcription termination/antitermination NusG family protein | - |
| PGC28_RS24755 (PGC28_24740) | 34347..35513 | - | 1167 | WP_000539529 | MobP1 family relaxase | - |
| PGC28_RS24760 (PGC28_24745) | 35516..36061 | - | 546 | WP_001436753 | DNA distortion polypeptide 1 | - |
| PGC28_RS24765 (PGC28_24750) | 36387..36659 | - | 273 | WP_000160399 | hypothetical protein | - |
| PGC28_RS24770 (PGC28_24755) | 36672..36821 | - | 150 | WP_000003880 | hypothetical protein | - |
| PGC28_RS24775 (PGC28_24760) | 36891..37070 | - | 180 | WP_000439078 | hypothetical protein | - |
| PGC28_RS24780 (PGC28_24765) | 37113..37442 | - | 330 | WP_000866649 | hypothetical protein | - |
| PGC28_RS24785 (PGC28_24770) | 37535..37750 | - | 216 | WP_001180118 | hypothetical protein | - |
| PGC28_RS24790 (PGC28_24775) | 37740..37985 | - | 246 | WP_000356542 | hypothetical protein | - |
Host bacterium
| ID | 9002 | GenBank | NZ_CP116043 |
| Plasmid name | pBL708 | Incompatibility group | IncX1 |
| Plasmid size | 40085 bp | Coordinate of oriT [Strand] | 35841..36447 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar 14[5]12:i:- strain BL708 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |