Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108524 |
| Name | oriT_p2S06B |
| Organism | Bradyrhizobium japonicum strain S06B-BJ |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP066353 (103370..103411 [+], 42 nt) |
| oriT length | 42 nt |
| IRs (inverted repeats) | 9..15, 20..26 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 42 nt
AAAGCAGGACGTCGCGGATGCGACGTATTACTGCGCCCTTGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file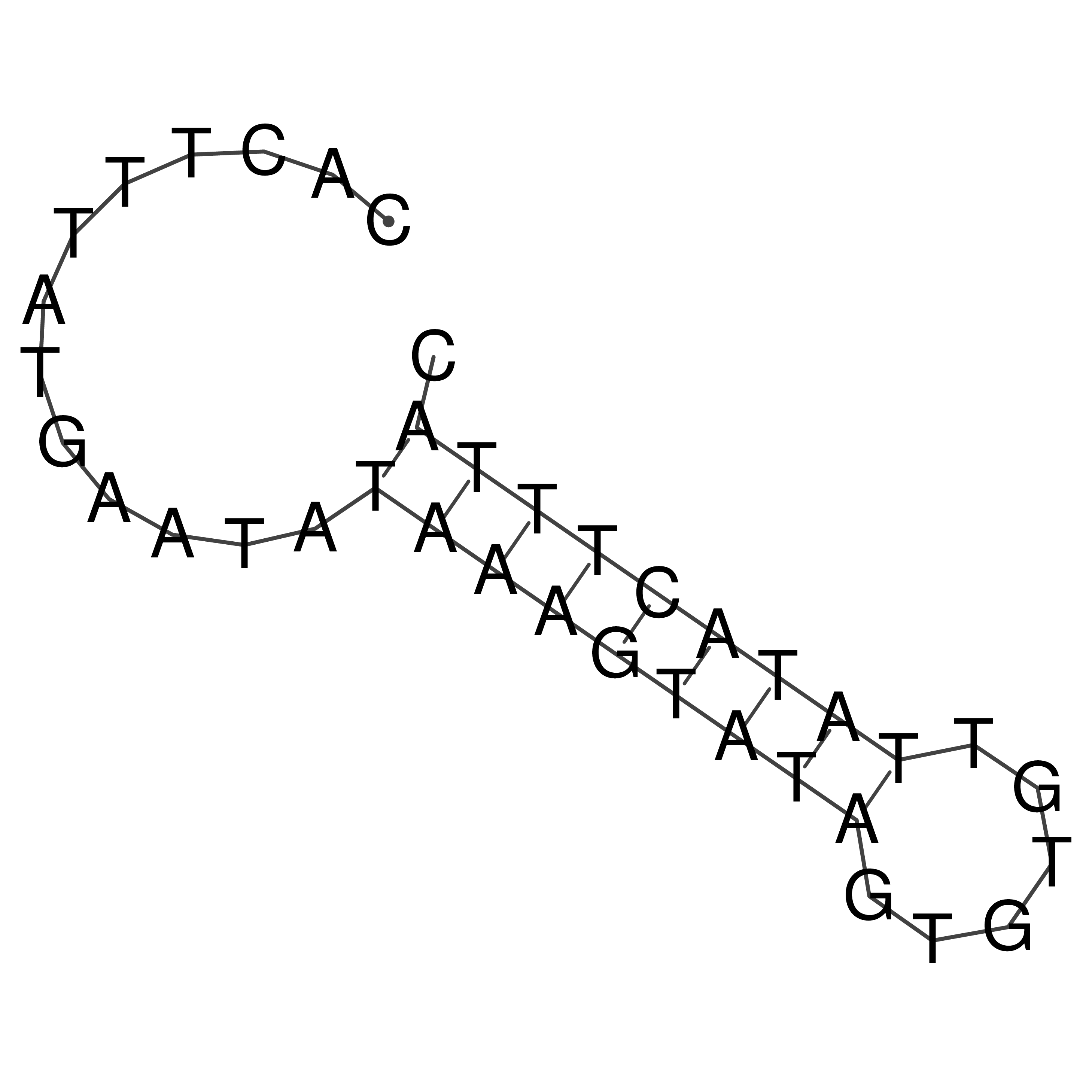
Relaxase
| ID | 5696 | GenBank | WP_212479306 |
| Name | traA_JEY30_RS50000_p2S06B |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 121918.26 Da Isoelectric Point: 9.9984
MAITHFTPQLISRGSGRSAVLSAAYRHCARMEHQAEGRTVDYSAKRGLRYEEFLLPPDAPQWARELIADR
SVAGAAEAFWNAIEAFEKRSDAQLAKEFIIALPVELTIDQNVALVREFVATQVLARGQVADWVFHDEPGN
PHIHLMTTLRPLGEDGFGPKKVPVIGHDGAPLRNAAGKIVYRLWSGEKAEFLAQREGWLDLQNRHLALAG
LDIRVDGRSYAERGVDLAPTTHIGVATKAIDRKGKKAGWSPKLERVALHEERKTDNRQRILRRPEIVLDL
VSREMSVFTERDIARVLHRYVDDAGTFQHLMARILQNPEALRLEGECIALATGIREPAKYTTRELIRLEA
EMAHRAVWLAGRSSHGVREKVLAAAFERHARLSEEQRVAIAHVASVGRIAAVVGRAGAGKTTMMKAAREV
WEAAGYRVVGAALAGKAAEGLEKEAGIASRTLASWELAWSNGREQLDDKTIFVLDEAGMVSSRQMALFVE
AVVKTGAKLALVGDPDQLQPIEAGAAFRAIAERTGYAELETIYRQREQWMRDASLDLARGRVAEALQAYG
ANGKLEARKYKAQAVEALIEDWNWDYDPAKSTLILAHLRRDVRVLNEMARAKLIERGIVEDGHAFRTEDG
PRSFATGDQIVFLKNEGSLGVKNGMIARVIEARPGWFAAMIGEGEARRRVEVDQRFYANVDHGYATTIHK
SQGATVDRVKVLATLSLDRHLAYVALTRHREDVALYYGAISFGKAGGLTKILSQSRAKETTLDYERGPSY
RAALRFAEARGLHLMRVARTLVADKVRWTVRQKQKLADLGARLLALGARLGLGAARKQTIPNGAKEAKPM
VAGITTFAKSIDQVVEDKLAADPGLKKQWEEVSTRFRFVFAEPESAFRKVDVDSMLKDKATAQSTLSKLA
EKPESFGALKGKTGLLASRADKQDRQRADLNVPALARDLERFLRLRSEAEHRYEAEERAARLKVAVDIPA
LSPSAKQTLERIRDAIDRNDLPAALEFALEDKMIEAELEGFARAVSERFGERSFVGLAAKDTDGKTFETL
SGGMNAAQKEGLKAAWPDLRTAQQLAAQQRTAVALKEAKTLRQTQSKGLSLQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 6073 | GenBank | WP_244430258 |
| Name | t4cp1_JEY30_RS49640_p2S06B |
UniProt ID | _ |
| Length | 353 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 353 a.a. Molecular weight: 38609.45 Da Isoelectric Point: 9.9739
MTPFANATTPVKAALGTALALGAVAVFLLVASNVLLLGLRLHDGGIHWSEIAKGWPRYGIDPRFDRWLTL
STLAGSIVVFGLLGAILRHRPLPLHGKARFASEREIKAAGLRSKDGLLLGRKDGAMLCFGGSEHVLLYAP
TRAGKGVGYVIPNLLNWPDSVVALDVKKENWDRSAGFRAAHGQEIHLFDPLDENGRTARYNPLGYIRTDP
ADLYDDLQRIAVMLFPAESRGDPFWFEAARSAFVAIGGYVAETPGLPLTIGETLRQLSATQDLKAHFDRI
IADRKAGPSPLSRHCITALNDFLAASENTLNSVRKTVTARLGLWLNPRIDAATSTNDFDLRQLRRRPMSI
YLG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 6074 | GenBank | WP_248890198 |
| Name | traG_JEY30_RS50015_p2S06B |
UniProt ID | _ |
| Length | 640 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 640 a.a. Molecular weight: 68705.10 Da Isoelectric Point: 9.9887
MTANRLLLLIVPMALMLGAIFGLTGMGHGLAGLGKTEAQRLLVARAGIAIPYIVAAAIGVVFLFAAAGSA
NIRTAAWSVVAGCAAAISMAAAREAIRLFAVPATAIPGRPPLVQIDPAIMVGAGIPLMAGIFALRVAVKG
NAAFTTPMPKRVRGRRALHGDADWMSMPEAARLFGIAGGIVVGERYRVDKDNAAAQPFRAMDRQSWGHGG
RAPLLCFDATFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVIKHRRAAGRTVHVLD
PKESGTGFNALDWIGQFGGRKEEDIAAVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEE
KNQTLRQVRANLSEPEPKLRARLQAIYDNSGSDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFVSDDLAGGTTDVFINLDLKTLETHAGLARVIIGSFMNAIYNRNGEVTGRALFLLDEVARLGF
MRILETARDAGRKYGITLLLLFQSIGQMRETYGGRDAASKWFESASWIAFAAVNDPETADYISKRCGMTT
VEIDQVSRSSQASGSSRTRSKQLASRPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRACVG
KNRFQPEEKT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 6075 | GenBank | WP_244430258 |
| Name | t4cp1_JEY30_RS50080_p2S06B |
UniProt ID | _ |
| Length | 353 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 353 a.a. Molecular weight: 38609.45 Da Isoelectric Point: 9.9739
MTPFANATTPVKAALGTALALGAVAVFLLVASNVLLLGLRLHDGGIHWSEIAKGWPRYGIDPRFDRWLTL
STLAGSIVVFGLLGAILRHRPLPLHGKARFASEREIKAAGLRSKDGLLLGRKDGAMLCFGGSEHVLLYAP
TRAGKGVGYVIPNLLNWPDSVVALDVKKENWDRSAGFRAAHGQEIHLFDPLDENGRTARYNPLGYIRTDP
ADLYDDLQRIAVMLFPAESRGDPFWFEAARSAFVAIGGYVAETPGLPLTIGETLRQLSATQDLKAHFDRI
IADRKAGPSPLSRHCITALNDFLAASENTLNSVRKTVTARLGLWLNPRIDAATSTNDFDLRQLRRRPMSI
YLG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 6076 | GenBank | WP_248890149 |
| Name | t4cp2_JEY30_RS50430_p2S06B |
UniProt ID | _ |
| Length | 392 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 392 a.a. Molecular weight: 43523.08 Da Isoelectric Point: 9.4312
MELESEIARSGNPDSSQKHHALKAHFDKIIANRKGGLSPLSRHCITALNDLLAASENTLNSVRKTVTARL
GLWLNPRIDAATLSNDFDLRQLRQQPMSIYLGVTPDNLDRMAPLLNLFFQQVVDLNTRELPEQNPKLTRK
VLLLLDEFPALGNVNVLAKSVAFRYGIRLLTVVQSPAQLRAIYGIDAARNFMTNHAVEVVFAPKEQDVAN
ELSERIGYETVNARSRSGPKGLAMRATSETVSEHRRALMLPQELKLLPKSKAFILGTGIPPIIADKIVYY
EDKAFLQRLLPAPVPELPKGRSNALLDAELKKLRSEIAELRAVFRARPMTDDEVADPSTIPANASFDFGD
VDIDLEGLSEDGMKAWTLNYIDAQAIPPARRSGPKREKRVAV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 8959 | GenBank | NZ_CP066353 |
| Plasmid name | p2S06B | Incompatibility group | - |
| Plasmid size | 279984 bp | Coordinate of oriT [Strand] | 103370..103411 [+] |
| Host baterium | Bradyrhizobium japonicum strain S06B-BJ |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |