Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108460 |
| Name | oriT_pSymB |
| Organism | Sinorhizobium meliloti strain S35m |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP065022 (750674..750708 [+], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 20..25, 30..35 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
>oriT_pSymB
GATCGGAGGGCGCAATATACGTCGCTGGCGCGACG
GATCGGAGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file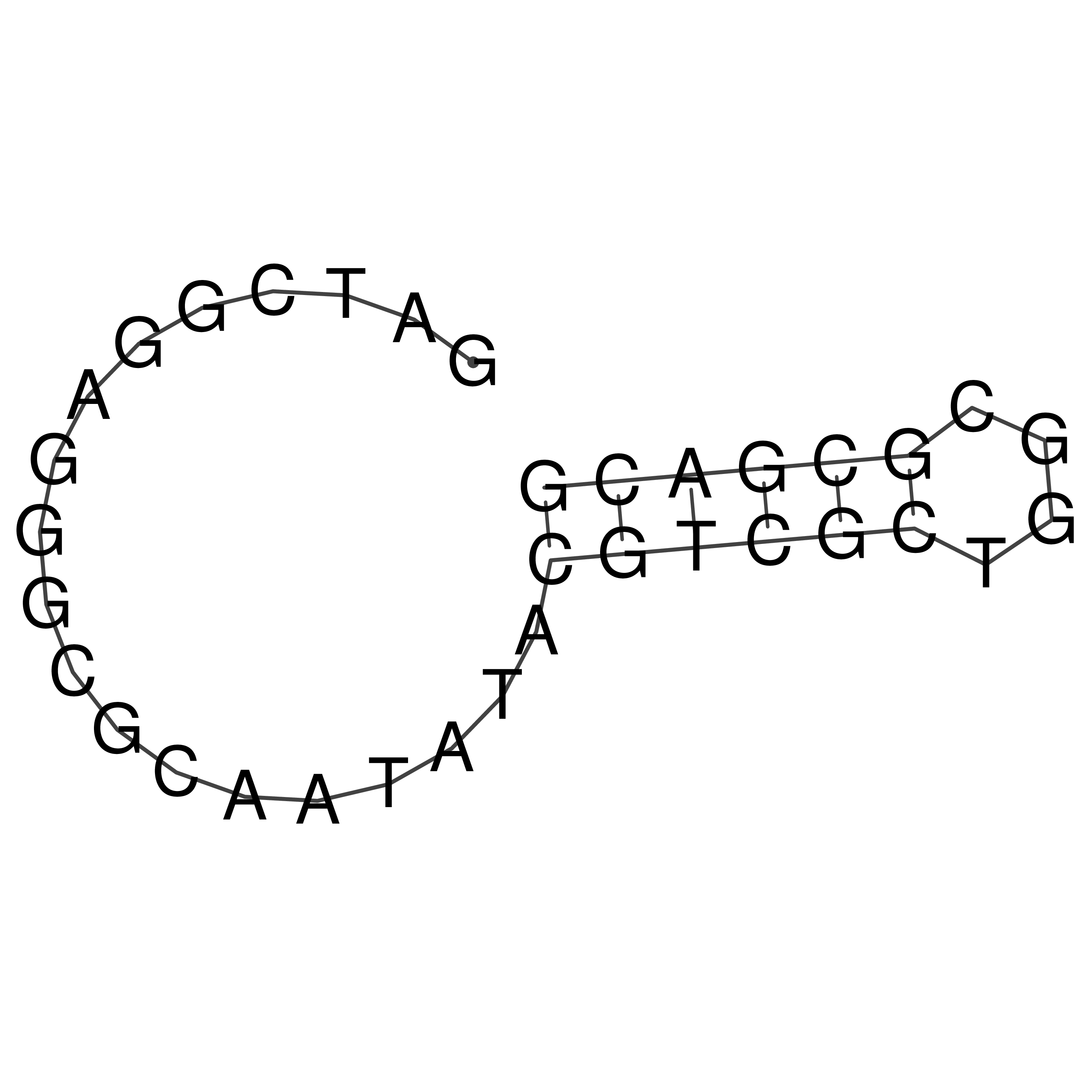
Relaxase
| ID | 5652 | GenBank | WP_206379526 |
| Name | traA_I0J99_RS27955_pSymB |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169680.23 Da Isoelectric Point: 7.6754
>WP_206379526.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEHGFGPKKVPVLGEGGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAEAGEAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGCEQLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRFFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRMLN
TALRNVMIGEGALAGAQEFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVEEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFAALVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKRELVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSERWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPRIAREFAKVSR
ALDERFGRNAILGGETDVINRVSPAQRRAFEAMRDRLTILQQAVRIQSSQKIVSERRQRAINQSRGIRM
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEHGFGPKKVPVLGEGGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAEAGEAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGCEQLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRFFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRMLN
TALRNVMIGEGALAGAQEFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVEEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFAALVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKRELVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSERWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPRIAREFAKVSR
ALDERFGRNAILGGETDVINRVSPAQRRAFEAMRDRLTILQQAVRIQSSQKIVSERRQRAINQSRGIRM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 6035 | GenBank | WP_127640223 |
| Name | tcpA_I0J99_RS31955_pSymB |
UniProt ID | _ |
| Length | 945 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 945 a.a. Molecular weight: 101440.16 Da Isoelectric Point: 4.5922
>WP_127640223.1 DNA translocase FtsK [Sinorhizobium meliloti]
MRIPRTNFSPAALYDANHADDYEDPHPAAQGAAHVPAAPEAAAHDHLFEAPEAPRRAPGHRDEETGYAGR
AARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPGWESHFFLSPNVRFTRTPERELMKRHPPAPEES
RIEADEAAAEASDAETVMDVVPAEPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSQAPEASVTKPAI
TESVAVAEEGPATSETTALPVTDEVPVVPHALPVANLAPESAAVEEDVSHQADARLAYLSDFAFFEFMPL
EVAAVPPTVTEPVKEAARIPAPIAAAPKVSPPKIVAAMPVEIRPQPPTAISSLFRVVECRRPEPATAEPV
TAAPQDIAAEPAAEAAALPAPRVVETPAPALAEPAVVPASESAPEAPVTRAAITMPAVIQRSSPSLPPIG
AIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEFE
PAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLALC
LGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGIPH
LLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGAPILATVQTGFEKGTGEP
LFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIKAN
FPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRPEY
LETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVER
MEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
MRIPRTNFSPAALYDANHADDYEDPHPAAQGAAHVPAAPEAAAHDHLFEAPEAPRRAPGHRDEETGYAGR
AARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPGWESHFFLSPNVRFTRTPERELMKRHPPAPEES
RIEADEAAAEASDAETVMDVVPAEPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSQAPEASVTKPAI
TESVAVAEEGPATSETTALPVTDEVPVVPHALPVANLAPESAAVEEDVSHQADARLAYLSDFAFFEFMPL
EVAAVPPTVTEPVKEAARIPAPIAAAPKVSPPKIVAAMPVEIRPQPPTAISSLFRVVECRRPEPATAEPV
TAAPQDIAAEPAAEAAALPAPRVVETPAPALAEPAVVPASESAPEAPVTRAAITMPAVIQRSSPSLPPIG
AIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEFE
PAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLALC
LGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGIPH
LLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGAPILATVQTGFEKGTGEP
LFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIKAN
FPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRPEY
LETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVER
MEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 8895 | GenBank | NZ_CP065022 |
| Plasmid name | pSymB | Incompatibility group | - |
| Plasmid size | 1716787 bp | Coordinate of oriT [Strand] | 750674..750708 [+] |
| Host baterium | Sinorhizobium meliloti strain S35m |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |