Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108405 |
| Name | oriT_p2-4 |
| Organism | Salmonella enterica strain |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP091574 (41310..41433 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTAGCTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file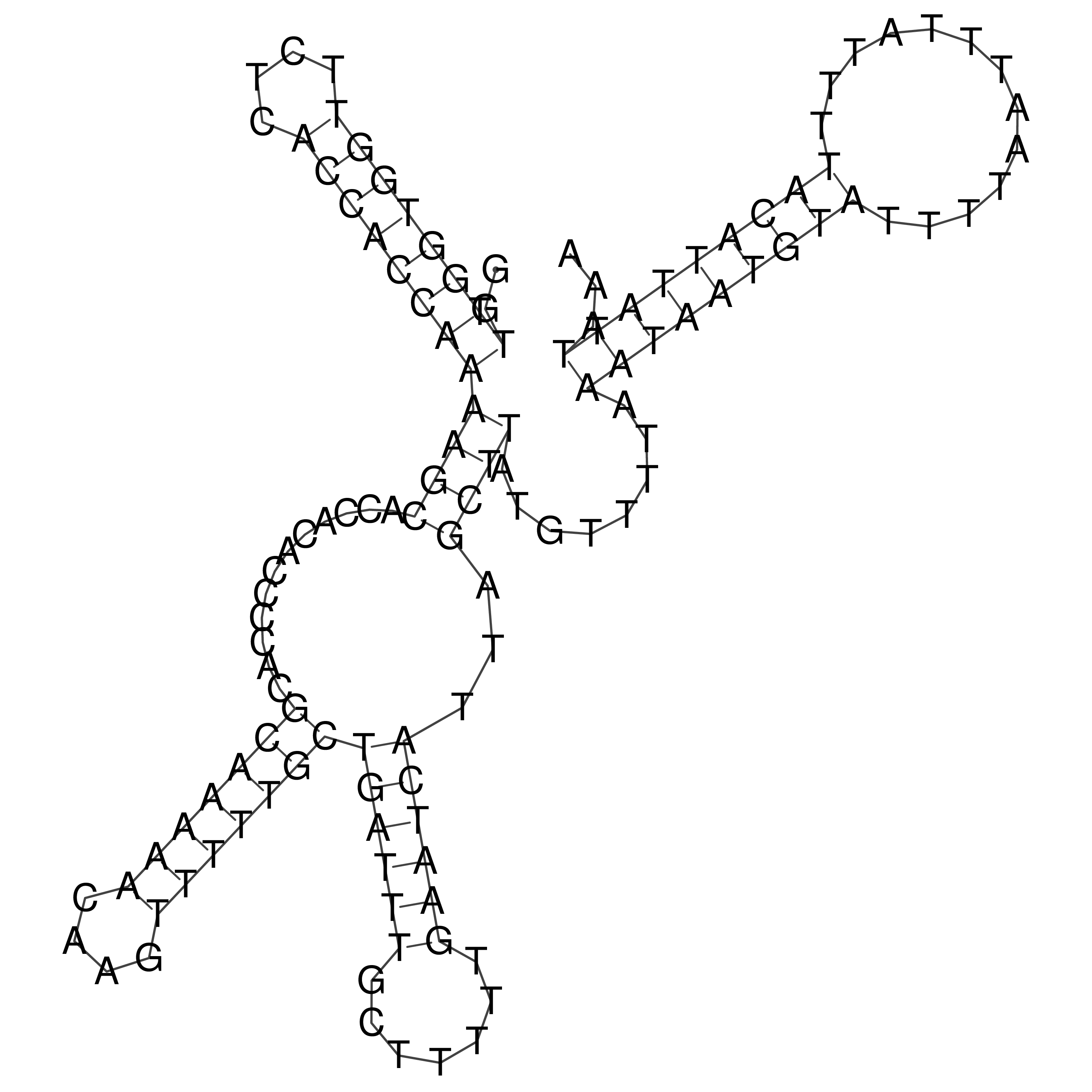
Relaxase
| ID | 5619 | GenBank | WP_247863749 |
| Name | traI_INN74_RS26090_p2-4 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192147.96 Da Isoelectric Point: 6.1283
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMR
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLPEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNIYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDVQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDVMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDKATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISHQKSALNTPAQQAIHLALPVVESKNLAFSHVDLLTEAKSFAAEGTSFADLGREIDA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETPDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISRDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPAQEQAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTSDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVREIAGQVRERAAITERETALPESVLREPQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 2704 | GenBank | WP_000124981 |
| Name | WP_000124981_p2-4 |
UniProt ID | A0A630FTW9 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14542.59 Da Isoelectric Point: 5.0278
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSVEMDRFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A630FTW9 |
| ID | 2705 | GenBank | WP_001254386 |
| Name | WP_001254386_p2-4 |
UniProt ID | A0A3Z6KJB5 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.11 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3Z6KJB5 |
T4CP
| ID | 6005 | GenBank | WP_015059012 |
| Name | traC_INN74_RS25980_p2-4 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99393.22 Da Isoelectric Point: 6.3465
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAAA
TQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPEMSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFKGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKKKQARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNRKVGEFIQKGYRTCRRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 6006 | GenBank | WP_086118634 |
| Name | traD_INN74_RS26085_p2-4 |
UniProt ID | _ |
| Length | 753 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 753 a.a. Molecular weight: 85673.46 Da Isoelectric Point: 5.0504
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPP
GISESGEVVDMAAYEAWQQENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 40738..68962
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| INN74_RS25875 (INN74_25875) | 36710..37144 | + | 435 | WP_000845953 | conjugation system SOS inhibitor PsiB | - |
| INN74_RS25880 (INN74_25880) | 37141..37860 | + | 720 | WP_001276217 | plasmid SOS inhibition protein A | - |
| INN74_RS25885 (INN74_25885) | 38082..38231 | + | 150 | Protein_48 | plasmid maintenance protein Mok | - |
| INN74_RS25890 (INN74_25890) | 38173..38298 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| INN74_RS25895 (INN74_25895) | 38617..38913 | - | 297 | Protein_50 | hypothetical protein | - |
| INN74_RS25900 (INN74_25900) | 39213..39509 | + | 297 | WP_001272251 | hypothetical protein | - |
| INN74_RS25905 (INN74_25905) | 39620..40441 | + | 822 | WP_001234445 | DUF932 domain-containing protein | - |
| INN74_RS25910 (INN74_25910) | 40738..41385 | - | 648 | WP_015059008 | transglycosylase SLT domain-containing protein | virB1 |
| INN74_RS25915 (INN74_25915) | 41662..42045 | + | 384 | WP_000124981 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| INN74_RS25920 (INN74_25920) | 42236..42922 | + | 687 | WP_015059009 | PAS domain-containing protein | - |
| INN74_RS25925 (INN74_25925) | 43016..43243 | + | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| INN74_RS25930 (INN74_25930) | 43277..43642 | + | 366 | WP_021519752 | type IV conjugative transfer system pilin TraA | - |
| INN74_RS25935 (INN74_25935) | 43657..43968 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| INN74_RS25940 (INN74_25940) | 43990..44556 | + | 567 | WP_000399794 | type IV conjugative transfer system protein TraE | traE |
| INN74_RS25945 (INN74_25945) | 44543..45271 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| INN74_RS25950 (INN74_25950) | 45271..46698 | + | 1428 | WP_032297072 | F-type conjugal transfer pilus assembly protein TraB | traB |
| INN74_RS25955 (INN74_25955) | 46688..47278 | + | 591 | WP_000002778 | conjugal transfer pilus-stabilizing protein TraP | - |
| INN74_RS25960 (INN74_25960) | 47265..47462 | + | 198 | WP_001324648 | conjugal transfer protein TrbD | - |
| INN74_RS25965 (INN74_25965) | 47474..47725 | + | 252 | WP_001038341 | conjugal transfer protein TrbG | - |
| INN74_RS25970 (INN74_25970) | 47722..48237 | + | 516 | WP_000809902 | type IV conjugative transfer system lipoprotein TraV | traV |
| INN74_RS25975 (INN74_25975) | 48372..48593 | + | 222 | WP_001278692 | conjugal transfer protein TraR | - |
| INN74_RS25980 (INN74_25980) | 48753..51380 | + | 2628 | WP_015059012 | type IV secretion system protein TraC | virb4 |
| INN74_RS25985 (INN74_25985) | 51377..51763 | + | 387 | WP_015059013 | type-F conjugative transfer system protein TrbI | - |
| INN74_RS25990 (INN74_25990) | 51760..52392 | + | 633 | WP_001203735 | type-F conjugative transfer system protein TraW | traW |
| INN74_RS25995 (INN74_25995) | 52389..53381 | + | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| INN74_RS26000 (INN74_26000) | 53408..53716 | + | 309 | WP_000412447 | hypothetical protein | - |
| INN74_RS26005 (INN74_26005) | 53779..54300 | + | 522 | WP_015059014 | hypothetical protein | - |
| INN74_RS26010 (INN74_26010) | 54327..54965 | + | 639 | WP_000087532 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| INN74_RS26015 (INN74_26015) | 54962..55333 | + | 372 | WP_000056336 | hypothetical protein | - |
| INN74_RS26020 (INN74_26020) | 55359..55781 | + | 423 | WP_001229309 | HNH endonuclease signature motif containing protein | - |
| INN74_RS26025 (INN74_26025) | 55778..57628 | + | 1851 | WP_015059015 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| INN74_RS26030 (INN74_26030) | 57655..57912 | + | 258 | WP_053913405 | conjugal transfer protein TrbE | - |
| INN74_RS26035 (INN74_26035) | 57905..58648 | + | 744 | WP_015059017 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| INN74_RS26040 (INN74_26040) | 58764..59045 | + | 282 | WP_000549793 | type-F conjugative transfer system pilin chaperone TraQ | - |
| INN74_RS26045 (INN74_26045) | 59032..59577 | + | 546 | WP_000059817 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| INN74_RS26050 (INN74_26050) | 59567..59881 | + | 315 | WP_001384255 | P-type conjugative transfer protein TrbJ | - |
| INN74_RS26055 (INN74_26055) | 59835..60227 | + | 393 | WP_000126338 | F-type conjugal transfer protein TrbF | - |
| INN74_RS26060 (INN74_26060) | 60214..61587 | + | 1374 | WP_000944325 | conjugal transfer pilus assembly protein TraH | traH |
| INN74_RS26065 (INN74_26065) | 61584..64400 | + | 2817 | WP_015059018 | conjugal transfer mating-pair stabilization protein TraG | traG |
| INN74_RS26070 (INN74_26070) | 64433..64954 | + | 522 | WP_000632668 | conjugal transfer entry exclusion protein TraS | - |
| INN74_RS26075 (INN74_26075) | 64979..65710 | + | 732 | WP_021559024 | conjugal transfer complement resistance protein TraT | - |
| INN74_RS26080 (INN74_26080) | 65913..66650 | + | 738 | WP_015059020 | hypothetical protein | - |
| INN74_RS26085 (INN74_26085) | 66701..68962 | + | 2262 | WP_086118634 | type IV conjugative transfer system coupling protein TraD | prgC |
Host bacterium
| ID | 8840 | GenBank | NZ_CP091574 |
| Plasmid name | p2-4 | Incompatibility group | IncFII |
| Plasmid size | 78621 bp | Coordinate of oriT [Strand] | 41310..41433 [+] |
| Host baterium | Salmonella enterica strain |
Cargo genes
| Drug resistance gene | fosA3, blaTEM-1B, blaCTX-M-55 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |