Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108336 |
| Name | oriT_pSD21_mcr8 |
| Organism | Enterobacter cloacae strain SD21 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP093916 (60563..60612 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pSD21_mcr8
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file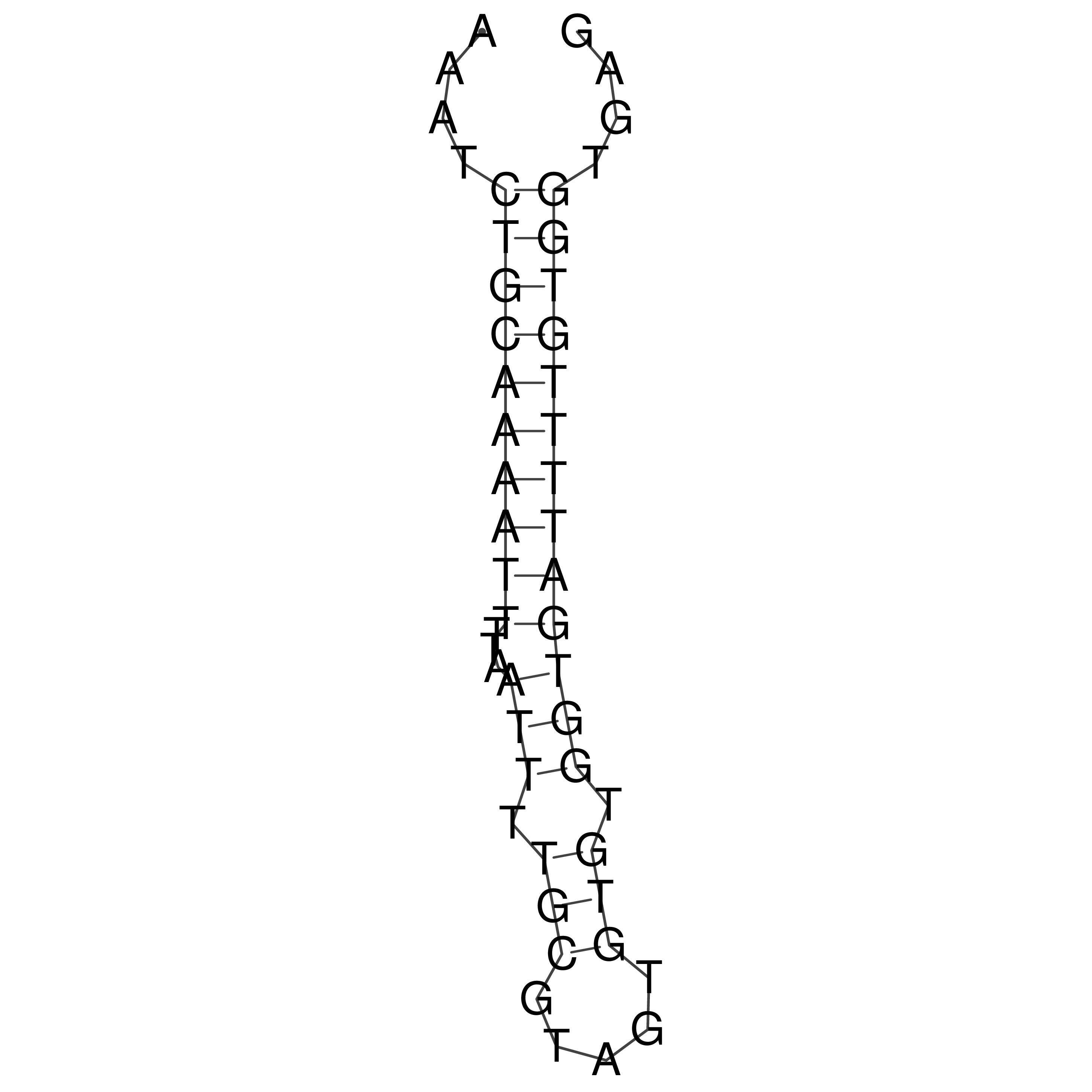
Relaxase
| ID | 5576 | GenBank | WP_022631527 |
| Name | traI_MQH03_RS24395_pSD21_mcr8 |
UniProt ID | U5N3A2 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190332.06 Da Isoelectric Point: 6.2397
>WP_022631527.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMSREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQLELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDTAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGGGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMSREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQLELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDTAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGGGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5952 | GenBank | WP_015065541 |
| Name | traD_MQH03_RS24390_pSD21_mcr8 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 86025.00 Da Isoelectric Point: 5.0577
>WP_015065541.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 60005..89687
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MQH03_RS24170 (MQH03_24170) | 56025..56381 | + | 357 | WP_019706019 | hypothetical protein | - |
| MQH03_RS24175 (MQH03_24175) | 56442..56654 | + | 213 | WP_019706020 | hypothetical protein | - |
| MQH03_RS24180 (MQH03_24180) | 56674..56889 | + | 216 | WP_142381705 | hypothetical protein | - |
| MQH03_RS24185 (MQH03_24185) | 56970..57290 | + | 321 | WP_015065521 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| MQH03_RS24190 (MQH03_24190) | 57280..57558 | + | 279 | WP_020801586 | helix-turn-helix transcriptional regulator | - |
| MQH03_RS24195 (MQH03_24195) | 57559..57960 | + | 402 | WP_015065522 | helix-turn-helix domain-containing protein | - |
| MQH03_RS24200 (MQH03_24200) | 58789..59610 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| MQH03_RS24205 (MQH03_24205) | 59643..59972 | + | 330 | WP_069335041 | DUF5983 family protein | - |
| MQH03_RS24210 (MQH03_24210) | 60005..60490 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| MQH03_RS24215 (MQH03_24215) | 60881..61297 | + | 417 | WP_072145360 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| MQH03_RS24220 (MQH03_24220) | 61497..62216 | + | 720 | WP_015065526 | hypothetical protein | - |
| MQH03_RS24225 (MQH03_24225) | 62349..62552 | + | 204 | WP_171486139 | TraY domain-containing protein | - |
| MQH03_RS24230 (MQH03_24230) | 62617..62985 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| MQH03_RS24235 (MQH03_24235) | 62999..63304 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| MQH03_RS24240 (MQH03_24240) | 63324..63890 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| MQH03_RS24245 (MQH03_24245) | 63877..64617 | + | 741 | WP_164530159 | type-F conjugative transfer system secretin TraK | traK |
| MQH03_RS24250 (MQH03_24250) | 64617..66041 | + | 1425 | WP_004194260 | F-type conjugal transfer pilus assembly protein TraB | traB |
| MQH03_RS24255 (MQH03_24255) | 66155..66739 | + | 585 | WP_015065529 | type IV conjugative transfer system lipoprotein TraV | traV |
| MQH03_RS24260 (MQH03_24260) | 66871..67281 | + | 411 | WP_032329942 | hypothetical protein | - |
| MQH03_RS24265 (MQH03_24265) | 67387..67605 | + | 219 | WP_004152501 | hypothetical protein | - |
| MQH03_RS24270 (MQH03_24270) | 67606..67917 | + | 312 | WP_004152502 | hypothetical protein | - |
| MQH03_RS24275 (MQH03_24275) | 67984..68388 | + | 405 | WP_004152503 | hypothetical protein | - |
| MQH03_RS24280 (MQH03_24280) | 68581..68823 | + | 243 | WP_226340492 | hypothetical protein | - |
| MQH03_RS24285 (MQH03_24285) | 68831..69229 | + | 399 | WP_011977783 | hypothetical protein | - |
| MQH03_RS24290 (MQH03_24290) | 69301..71940 | + | 2640 | WP_004152505 | type IV secretion system protein TraC | virb4 |
| MQH03_RS24295 (MQH03_24295) | 71940..72329 | + | 390 | WP_004152506 | type-F conjugative transfer system protein TrbI | - |
| MQH03_RS24300 (MQH03_24300) | 72329..72955 | + | 627 | WP_009309871 | type-F conjugative transfer system protein TraW | traW |
| MQH03_RS24305 (MQH03_24305) | 72997..73404 | + | 408 | WP_248766918 | hypothetical protein | - |
| MQH03_RS24315 (MQH03_24315) | 74160..75149 | + | 990 | WP_009309872 | conjugal transfer pilus assembly protein TraU | traU |
| MQH03_RS24320 (MQH03_24320) | 75162..75800 | + | 639 | WP_164530161 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| MQH03_RS24325 (MQH03_24325) | 75859..77814 | + | 1956 | WP_013023827 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| MQH03_RS24330 (MQH03_24330) | 77846..78100 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| MQH03_RS24335 (MQH03_24335) | 78078..78326 | + | 249 | WP_004152675 | hypothetical protein | - |
| MQH03_RS24340 (MQH03_24340) | 78339..78665 | + | 327 | WP_004152676 | hypothetical protein | - |
| MQH03_RS24345 (MQH03_24345) | 78686..79438 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| MQH03_RS24350 (MQH03_24350) | 79449..79688 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| MQH03_RS24355 (MQH03_24355) | 79660..80217 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| MQH03_RS24360 (MQH03_24360) | 80263..80706 | + | 444 | WP_004194305 | F-type conjugal transfer protein TrbF | - |
| MQH03_RS24365 (MQH03_24365) | 80684..82063 | + | 1380 | WP_164530163 | conjugal transfer pilus assembly protein TraH | traH |
| MQH03_RS24370 (MQH03_24370) | 82063..84912 | + | 2850 | WP_013609534 | conjugal transfer mating-pair stabilization protein TraG | traG |
| MQH03_RS24375 (MQH03_24375) | 84915..85448 | + | 534 | WP_014343486 | conjugal transfer protein TraS | - |
| MQH03_RS24380 (MQH03_24380) | 85634..86365 | + | 732 | WP_032329926 | conjugal transfer complement resistance protein TraT | - |
| MQH03_RS24385 (MQH03_24385) | 86557..87246 | + | 690 | WP_015065540 | hypothetical protein | - |
| MQH03_RS24390 (MQH03_24390) | 87375..89687 | + | 2313 | WP_015065541 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 8771 | GenBank | NZ_CP093916 |
| Plasmid name | pSD21_mcr8 | Incompatibility group | IncFIA |
| Plasmid size | 100852 bp | Coordinate of oriT [Strand] | 60563..60612 [-] |
| Host baterium | Enterobacter cloacae strain SD21 |
Cargo genes
| Drug resistance gene | mcr-8 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |