Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108333 |
| Name | oriT_p116-2 |
| Organism | Enterococcus faecium strain 116 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP047329 (10784..10813 [-], 30 nt) |
| oriT length | 30 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 30 nt
>oriT_p116-2
AACGTAAGTTAGTGGGTATAAGTGCGCCCT
AACGTAAGTTAGTGGGTATAAGTGCGCCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file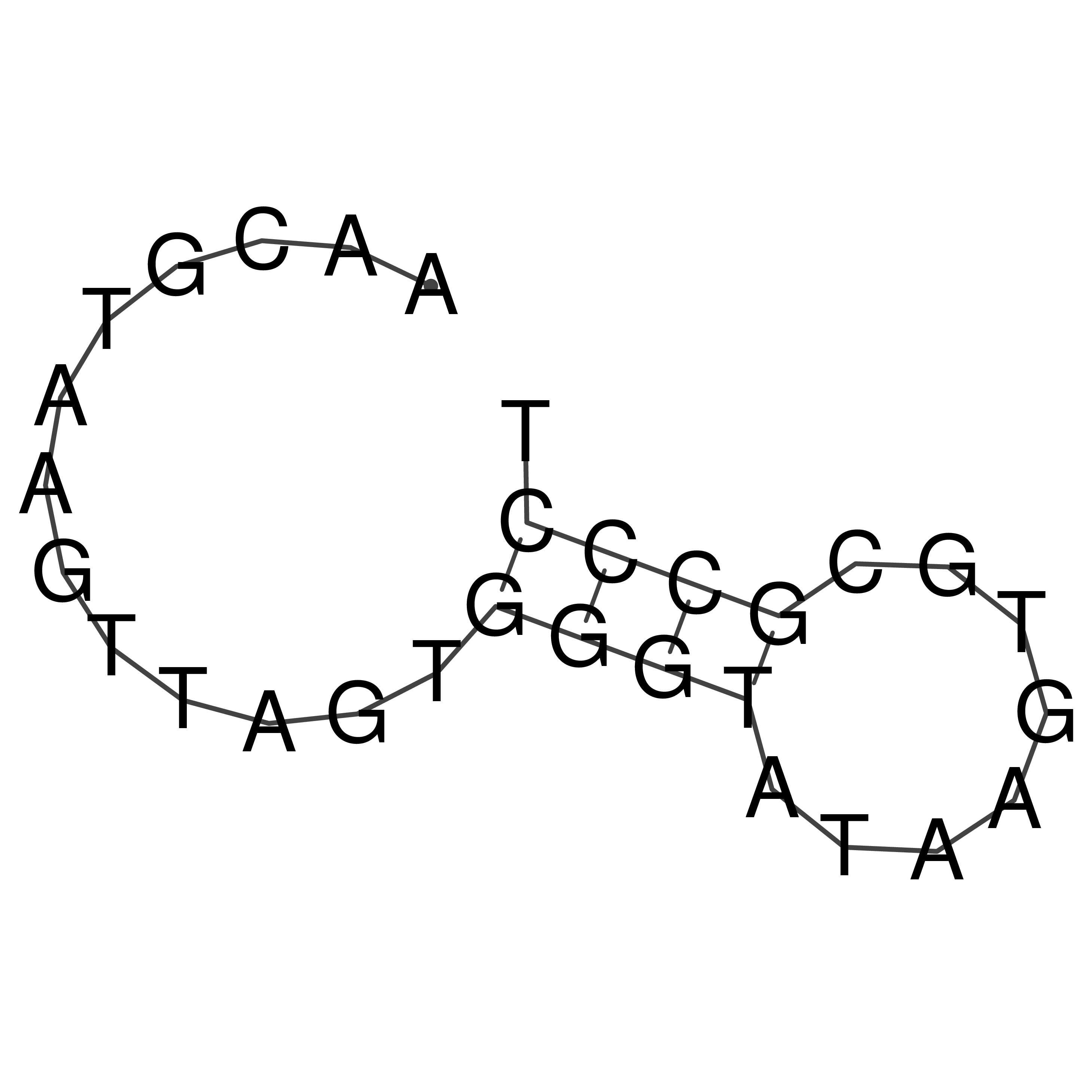
Relaxase
| ID | 5574 | GenBank | WP_242693023 |
| Name | mobQ_GST81_RS13120_p116-2 |
UniProt ID | _ |
| Length | 664 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 664 a.a. Molecular weight: 77704.92 Da Isoelectric Point: 9.3434
>WP_242693023.1 MULTISPECIES: MobQ family relaxase [Enterococcus]
MIQLAIFHLSMTIAKREGGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTIKPEAFILKPDYVPDEFLDR
QTLWNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHI
MLTMREVDSEGNILNKRKRIPKLDENGNQIFNEKGQRVTVSIKTNDWDRKSLVSEIRKDWADKVNQYLKD
RNIDQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLESQEKVLKQ
DQNFTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISS
ERDMLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVIEKEE
SKKIFKEKPFQTSRYLDSKIKQVEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVS
IDSVIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMK
YIDELSPLAKHNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRL
LSGNQLQKKRNLDKLIKQTKAKKNQSLQRNIPLR
MIQLAIFHLSMTIAKREGGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTIKPEAFILKPDYVPDEFLDR
QTLWNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHI
MLTMREVDSEGNILNKRKRIPKLDENGNQIFNEKGQRVTVSIKTNDWDRKSLVSEIRKDWADKVNQYLKD
RNIDQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLESQEKVLKQ
DQNFTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISS
ERDMLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVIEKEE
SKKIFKEKPFQTSRYLDSKIKQVEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVS
IDSVIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMK
YIDELSPLAKHNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRL
LSGNQLQKKRNLDKLIKQTKAKKNQSLQRNIPLR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5950 | GenBank | WP_002325630 |
| Name | t4cp2_GST81_RS13165_p116-2 |
UniProt ID | _ |
| Length | 551 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 551 a.a. Molecular weight: 63114.14 Da Isoelectric Point: 4.7040
>WP_002325630.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Bacilli]
MSSLLAESDGLILGKFQSGNVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVAEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELLMSDILSSFDEGIEENTTNNDNSELPF
MSSLLAESDGLILGKFQSGNVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVAEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELLMSDILSSFDEGIEENTTNNDNSELPF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13279..25605
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GST81_RS13100 (GST81_13010) | 9215..9712 | + | 498 | WP_071621495 | DnaJ domain-containing protein | - |
| GST81_RS13105 (GST81_13015) | 9746..10051 | + | 306 | WP_033626036 | hypothetical protein | - |
| GST81_RS13110 (GST81_13020) | 10058..10327 | - | 270 | WP_212481480 | hypothetical protein | - |
| GST81_RS13470 | 10490..10627 | - | 138 | WP_249222588 | hypothetical protein | - |
| GST81_RS13120 (GST81_13030) | 10919..12904 | + | 1986 | WP_165548714 | MobQ family relaxase | - |
| GST81_RS13125 (GST81_13035) | 12928..13260 | + | 333 | WP_000713503 | CagC family type IV secretion system protein | - |
| GST81_RS13130 (GST81_13040) | 13279..13662 | + | 384 | WP_165548716 | glycosyltransferase | trsC |
| GST81_RS13135 (GST81_13045) | 13679..14308 | + | 630 | WP_002301630 | hypothetical protein | trsD |
| GST81_RS13140 (GST81_13050) | 14319..16280 | + | 1962 | WP_165548718 | TrsE protein | virb4 |
| GST81_RS13145 (GST81_13055) | 16294..17646 | + | 1353 | WP_104659242 | conjugal transfer protein TraF | trsF |
| GST81_RS13150 (GST81_13060) | 17668..18777 | + | 1110 | WP_021109240 | lysozyme family protein | orf14 |
| GST81_RS13155 (GST81_13065) | 18790..19341 | + | 552 | WP_021109241 | hypothetical protein | - |
| GST81_RS13160 (GST81_13070) | 19346..19777 | + | 432 | WP_002301637 | hypothetical protein | - |
| GST81_RS13165 (GST81_13075) | 19770..21425 | + | 1656 | WP_002325630 | VirD4-like conjugal transfer protein, CD1115 family | - |
| GST81_RS13170 (GST81_13080) | 21443..22366 | + | 924 | WP_212481474 | hypothetical protein | - |
| GST81_RS13175 (GST81_13085) | 22368..23300 | + | 933 | WP_002325632 | conjugal transfer protein TrbL family protein | trsL |
| GST81_RS13180 (GST81_13090) | 23317..24288 | + | 972 | WP_002425009 | hypothetical protein | - |
| GST81_RS13185 (GST81_13095) | 24317..24685 | + | 369 | WP_000516429 | hypothetical protein | - |
| GST81_RS13190 (GST81_13100) | 24745..25605 | + | 861 | WP_212481476 | LPXTG cell wall anchor domain-containing protein | prgC |
| GST81_RS13195 (GST81_13105) | 25928..26200 | + | 273 | WP_011266100 | helix-turn-helix transcriptional regulator | - |
| GST81_RS13200 (GST81_13110) | 26584..28074 | + | 1491 | WP_212481478 | primase C-terminal domain-containing protein | - |
| GST81_RS13205 | 28423..28593 | + | 171 | WP_000713594 | hypothetical protein | - |
| GST81_RS13210 (GST81_13115) | 28607..29224 | + | 618 | WP_001062589 | recombinase family protein | - |
Host bacterium
| ID | 8768 | GenBank | NZ_CP047329 |
| Plasmid name | p116-2 | Incompatibility group | - |
| Plasmid size | 31694 bp | Coordinate of oriT [Strand] | 10784..10813 [-] |
| Host baterium | Enterococcus faecium strain 116 |
Cargo genes
| Drug resistance gene | fosB3, erm(B), aac(6')-aph(2'') |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |