Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108332 |
| Name | oriT_pCB3126_2 |
| Organism | Sinorhizobium terangae strain CB3126 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP121661 (545152..545186 [+], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 20..25, 30..35 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
ATCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file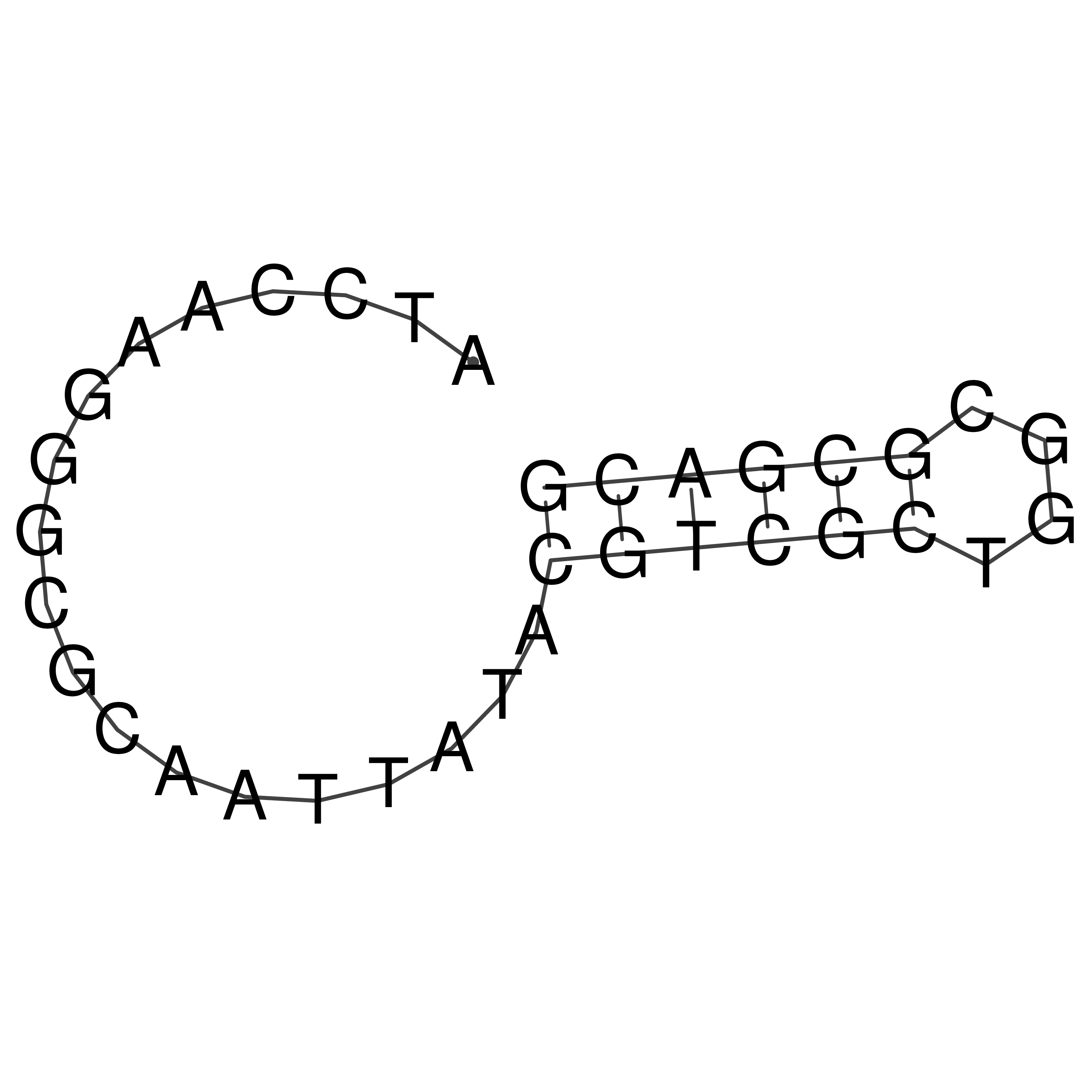
Relaxase
| ID | 5573 | GenBank | WP_283067698 |
| Name | traA_QA637_RS30755_pCB3126_2 |
UniProt ID | _ |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122341.81 Da Isoelectric Point: 9.4340
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYRAKQGLLHEEFVIPADAPQWLRAMIADH
SVSGASEAFWNKVEVFEKRSDAQLAKDITIALPIELSTEQSIALMRDFVERHITSNGMVADWVYHDAPDN
PHVHLMTTLRPLTEDGFGAKKVAVVGPDGNPIRNDAGKIVYELWAGSADDFNAFRDGWFACQNKHLALAG
LDIRVDGRSFERQGIELAPTIHLGVGAKAIERKSAEEDRDISLERLELQEERRAENARRIQRRPEIVLDL
ITREKSVFDERDVAKILHRYIGDAGLFQSLMARILQSPETLRLERERIEFSSGTRAPAKYTTRELIRLEA
RMANRALWLSQRSSHDVRDAVLDATFACHSRLSEEQTSAIAHVTGGERIAAVIGRAGAGKTTMMKAARET
WEAAGYRVLGGALAGKAAEGLQREAGILSRTLSSWELRWKEGRNQLDDRTVFVLDEAGMVSSRQMALFVE
AAVNAGAKLVLIGDPEQLQPIEAGAAFRGIADRIGYAELETIYRQRQQWMRDASLDLARGNVRNAVEAYH
AQGHVNGSELKADAVRALIADWNRDYDPSKSSLILAHLRRDVRMLNEMARAKLVERGIVADGFAFGTEDG
DRKFAAGDQIVFLKNEGSLGVKNGMLAKVVNAAPGRIVAEVGEGEHRRQVTVEQRFYSHVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLSYVAMTRHRDDLGLYYGRLSVAKAGGLIPILSRKNAKETTLDYVHGSFY
RQALRFAEARGLHLVNVARTVVRDRLDWTVRQKSKLIELGARLAMIAGKLGLSAATVRTFQSHTIKESKP
MVSGITTFPNSLEQDVEDRLAADPGLKKQWEEVSARFHLVYAEPQAAFKTVNVDAILKDEAAAKSTIARI
ASEPESFGALNGKTGILASRADKQSRETAVTNAPALSRNLERYLRQRTEAERKYEAEERTVRLKLSLDIP
ALSSAAKQTLEHVRDAIDRNDLPAGLEYALSDKMVKAELEGFARAVSERFGERIFLPLAAKDTNGQAFQF
VTSGMSAGQKAEVQSAWNSMRTVQQLSAHQRTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5947 | GenBank | WP_283067739 |
| Name | tcpA_QA637_RS28505_pCB3126_2 |
UniProt ID | _ |
| Length | 1751 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 1751 a.a. Molecular weight: 194714.74 Da Isoelectric Point: 5.2837
MDIHELIGKVACSILKEELGDVGGEAGTARFLLDGLSIPQTLAVTRAVLADVFLSERVDIKLPRTLFDGH
ALPEEILTDRNATFYRSADCEKAAFLITNASADEGQAEDMSLHEVTPIGAAQLIERLPAWVSAASAGLAL
TDDAKAWWERSLAGLSQVGSVALERFARYIVATREAVIDEGYPIIEALGYALPALQLPRDPAAFAGIKDK
SRRHASVWKREYIGLRRKRHPYLLKQNPNQIVISESELRDAFEKARDVIPEIVHPVVELFIESRPAWNSS
SEALANCLWEHVKPLFEGLAREKTNLGQDTQRFYSEGPEDLLSKDDEEHLELLARRKTTASPEPEDIDFY
ERHRDEIREDRKLKSSWDKFIYGRPLETEDFLSGVAMMMDTLNARAAVGVERRLTIRCDSLTKRDLRSLN
TEAGLFFSLRYAGLPKLLGTNVAIEFGSLMDYPAVLRGWQDAKDKSAINRSTAKAALQLRFQLELETTDF
EGGTSIASAQLIWKYRSDVISSQLSDDWDRLCSNAFVALQCGREPGMAGRRPGSIDLSDVKTLVPAYDRD
RGSLVPTYTRERDLRLIWQANLKAALEQDLIDAAVSTTLSASFESFCERYQTAIRSFRTEGASNPVCRDQ
AASYAELLDLVRTLAPGDRNKDLLLRPLLELGQAPVGDGAAAAIVAPWHPLRLAAIWRKAHLVREVVRKI
IDLPAGLDGDTKLFFRDLAEDIRHVFYPEVVVSWSGRKPELLSLADHQGDYSLHERPVLDGAGGGETNDD
AKAGTNCLLDLTQRYLNLHPHERANMSLVLYNCDSARLPQQLVDGLGELNDDEDMRCQVMLRHTDGTRLR
DIYRSILTSTSGSAEVLAASEITQDFMARLRISVIADQAPPPDPRDGRPYDIVFSQDVISRHASVEWYRE
SADPADLATLLPARWSRRRPGAMDDLKSSVYLCSPVQSREGWAFLSATTAFLKNDEDLQDGKRLLPVRQL
DFRDDRTARIFEETHDLGAWVVNFDELLDRRQLQNQSVRVIRYKQSATQGRNVVISSRAPLTLLRAMVRR
RLEELQLDLSPEDINALADRFIGDANDISGDIVLRAAKRGEAAGELIGVVLSRFLARQSLGADRLVGWYF
LDDYASWLGAREETQADILALSPAIDASGQLTLRIIVTEAKYVDAVSFPSKRKESEKQLRDTMRRIIDAT
FEPDRLDRESWLSRLGDLLLDGIQIPAASNVPLVDWRRRIRDGLARIEVVGLSHVFVPTALDGSTVTDFS
EVSGAPASRQMIFGRAELKDLITAYIANANVEALLTSIDPIYGKVPAPHTPGQTINPRSDAPRAVAKAKS
EPVAENDDRRSAPPSPAEKPSDENALKPEVTNTIAPASSVAEIVERHLDTSPRQTEADEEWLANTAMMAR
SALQQLGLQAKLIDQKLTPNAAILRFSGSANLTVEQVLKRRSELLTTHRLDIISVRPEPGAIAIYVARTQ
RQIVDIQSLWSRWKPTISPTGNQNLVIGVREDDNELFILSPSQRHAPHTLIAGSTGSGKSVLMQSIILGI
AATNDPRHAKIVLIDPKQGVDYFAFDALPHLEDGIIDQQETAIDQLERMVLEMDRRYRTFKEARVANITA
YNAKATPAEVLPTYWVIHDEFAEWMLTDDYRVAVTSTVGRLGVKARAAGIHLIFAAQRPEANVMPMQLRS
QLGNRLILRVDSEGTSEIALGERGAERLLGKGHMIVRAEGEQSLIYAQVPFASEGFVSDVVAAIRGRGDQ
Q
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5948 | GenBank | WP_283067694 |
| Name | traG_QA637_RS30740_pCB3126_2 |
UniProt ID | _ |
| Length | 640 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 640 a.a. Molecular weight: 69598.97 Da Isoelectric Point: 9.0656
MTRIVLFLAPCALMLLVIIGMTGTENWLPVFGKSEAAQQTLGRAGIALPYLVAALLGIVFMFASAGSARI
RTAGWGVLAGATATILIAAMREFVRLSAFAGQVPGGKSIFSYVDPATTIGVCAALMSALFGLRVVIAGNA
AFAKAEPKRIYGKRALYGEAEWMKLPEAEKLFPERGGIVIGERYCVDKDSVAAQSFRADSAETWGAGGKA
PLLCFDGSFGPSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVSKHRGGANRDVFVLDPK
NSEIGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTREPE
QTLRQVRKNLSEPEPKLRERLQSIYHNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYLNYAAL
VSGTTFTTSDIAEGNTDVFINIDLKTLETHAGLARVIIGSFLNAIYNRNGEMEGRALFLLDEVARLGYMR
ILETARDAGRKYGIMLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSFQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRREDMKACVKLN
VLFRASAEKR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5949 | GenBank | WP_283067713 |
| Name | t4cp2_QA637_RS30830_pCB3126_2 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91866.57 Da Isoelectric Point: 6.1105
MVALRRFRATGPSFADLVPYAGLVGNGVLLLKDGSLMAGWYFAGPDSESATALERNELSRQINAILSRLG
SGWMIQVEAIRFPTIDYPSEDRCHFPDPVTRAIDAERRAHFEREQGHFESKHALILTYRPLESKRTALSK
YIYSDEESRKKSYADTVLFMFKNAVREIEQYMANSLSIRRMETHEVSEREGTRVARYDELLQFVRFCITG
HSHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWLQKVRPFFDQLFQTQSRSIDQDAMTMVTETEDAIAHASSQLVAYGYYTPVIVL
FDNDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYSNIREPLINTSNLADLIPLNAVWS
GNPAAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGNSLLPLTLAGDGDHYEIGGDNEGEGRALAFCPLSALGNDADRAWATEWIEMLVALQGVTITPDHRN
AISRQVELMANARGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLSLGALQTFEIEHLMNMG
ERNLVPVLTYLFRRIERRLDGSPSLIVLDEAWLMLGHPVFRAKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGLNERQIDIVASATPKREYYATTPEGRRLFDMALGP
VALSFVGASGKDELKRIRALKSEHGRDWPIHWLNTRGVHDAASLLNFE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 555746..565082
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QA637_RS30775 (QA637_30775) | 551339..552043 | - | 705 | WP_283067701 | autoinducer binding domain-containing protein | - |
| QA637_RS30780 (QA637_30780) | 552407..553660 | + | 1254 | WP_283067702 | ATP-binding protein | - |
| QA637_RS30785 (QA637_30785) | 553982..554188 | - | 207 | WP_283067703 | helix-turn-helix transcriptional regulator | - |
| QA637_RS30790 (QA637_30790) | 554389..554709 | + | 321 | WP_283067704 | transcriptional repressor TraM | - |
| QA637_RS30795 (QA637_30795) | 554713..555417 | - | 705 | WP_283067706 | autoinducer binding domain-containing protein | - |
| QA637_RS30800 (QA637_30800) | 555746..557047 | - | 1302 | WP_283067708 | IncP-type conjugal transfer protein TrbI | virB10 |
| QA637_RS30805 (QA637_30805) | 557059..557496 | - | 438 | WP_283067710 | conjugal transfer protein TrbH | - |
| QA637_RS30810 (QA637_30810) | 557499..558311 | - | 813 | WP_283067711 | P-type conjugative transfer protein TrbG | virB9 |
| QA637_RS30815 (QA637_30815) | 558327..558989 | - | 663 | WP_283067712 | conjugal transfer protein TrbF | virB8 |
| QA637_RS30820 (QA637_30820) | 559014..560162 | - | 1149 | WP_283068068 | P-type conjugative transfer protein TrbL | virB6 |
| QA637_RS30825 (QA637_30825) | 560198..561001 | - | 804 | WP_283068069 | P-type conjugative transfer protein TrbJ | virB5 |
| QA637_RS30830 (QA637_30830) | 560973..563429 | - | 2457 | WP_283067713 | conjugal transfer protein TrbE | virb4 |
| QA637_RS30835 (QA637_30835) | 563440..563736 | - | 297 | WP_283067715 | conjugal transfer protein TrbD | virB3 |
| QA637_RS30840 (QA637_30840) | 563729..564115 | - | 387 | WP_283067717 | TrbC/VirB2 family protein | virB2 |
| QA637_RS30845 (QA637_30845) | 564105..565082 | - | 978 | WP_283067719 | P-type conjugative transfer ATPase TrbB | virB11 |
| QA637_RS30850 (QA637_30850) | 565093..565716 | - | 624 | WP_283067720 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 8767 | GenBank | NZ_CP121661 |
| Plasmid name | pCB3126_2 | Incompatibility group | - |
| Plasmid size | 566092 bp | Coordinate of oriT [Strand] | 545152..545186 [+] |
| Host baterium | Sinorhizobium terangae strain CB3126 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | linJ, dxnH, LinG |
| Symbiosis gene | nodD, nodM, nifX, mLTONO_5203, nifN, nifE, nifD, nifH, nifQ, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nodH, nodJ, nodI, nodU, nodS, nodB, nodA |
| Anti-CRISPR | - |