Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108327 |
| Name | oriT_pCC283b_5 |
| Organism | Rhizobium leguminosarum strain CC283b |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP121640 (107343..107392 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pCC283b_5
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTGCTGAAGGG
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTGCTGAAGGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file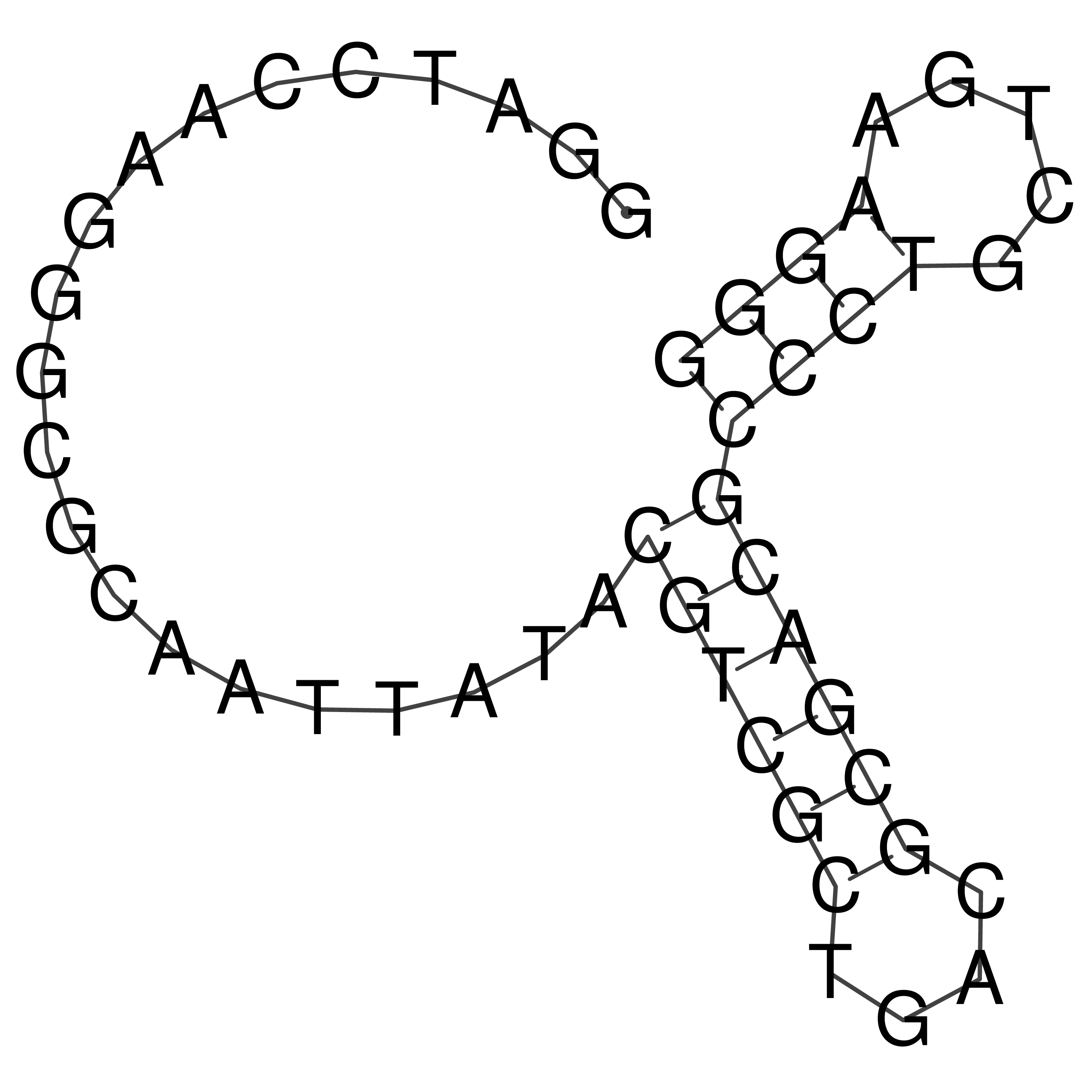
Relaxase
| ID | 5568 | GenBank | WP_027690605 |
| Name | traA_QA638_RS39070_pCC283b_5 |
UniProt ID | _ |
| Length | 1197 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1197 a.a. Molecular weight: 131914.80 Da Isoelectric Point: 9.2524
>WP_027690605.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYSRKQGLLHEEFVIPETAPDWLRSMIADR
SVSGASEAFWNKVEAFEKRADAQLAKDVTIALPVELSNDQNIALVRDFVERHITAKGMVADWVYHDALGN
PHIHLMTTLRPLTEDGFGAKKVAVLAPDGKPVRNDAGKIVYELWAGSTDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKAASGNGLAEEKVALERLELQEERRAENARRIQRNPEI
VVDLITREKSVFDERDIAKVLYRYIDDAALFQNLMARLLQSPQTLRLDRERMDLVTGVRAPAKYTTRELI
RLEAQMANQAIWLSQRSSHRVNRTVLSGIVSRHDRLSDEQKTAIEHVAGPERIAAVIGRAGAGKTTMMKA
AREAWEAAGYRVVGGALAGKAADGLEKEAGIASRTLSAWELRWDQERDRLDEKSVFVLDEAGMVSSRQMA
RFVEAVTLSGAKLVLVGDPEQLQPIEAGAAFRAIAERIGYAELETIYRQREQWMRDASLDLARGNVSAAL
DAYAQRDLVRTGWTRDEAITALIAEWDHEYDPAKSTLILAHRRVDVRLLNEMARSKLVERGLIEAGHAFK
TEDGTRQFAAGDQIVFLKNEGSLGVKNGMLALVVDAQPGRIVAEIGNGEDRRRVVVEQRFYANVDHGYAT
TVHKSQGATVDRVKVLASSTLDRHLSYVAMTRHRETAELYVGLEEFAQRRGGVLIAHGEAPYEHKPGNRD
SYYVTLGFANGQERTVWGVDLARAMDASDSRIGDRIGLKHVGSQRVTLPNGTEVDRNSWKVVPIQELAMA
RLHERLSRAGSKETTLDYQDASHYRAALRFAEARGLHLMNVARTIAHDQLQWTVRQSSKLAELGARLVAV
AAKLGLGGAKSTVSTISVIKEAKPMVFGTTTFPRSIGQAVEDKLSADPGLKASWQEVSARFHHVFADPQA
AFKAVNVDAMLANGTVAATTIVRIAEQPESFGALKGKTGLFAGSAEKQARDTALVNAPALARDLQGFIAK
RAAAARRYEDEERAVRAQLSLDIPALSASGKQVLERVRDAIDRNDIPAGLEFALADKMVKAELEGFAKAV
SERFGERTFLPLAAKTADGKAFEVASAGMQPAQKNELRSAWDTIRTVQQLAAHERTAVALKQAEAIRQTQ
TKGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYSRKQGLLHEEFVIPETAPDWLRSMIADR
SVSGASEAFWNKVEAFEKRADAQLAKDVTIALPVELSNDQNIALVRDFVERHITAKGMVADWVYHDALGN
PHIHLMTTLRPLTEDGFGAKKVAVLAPDGKPVRNDAGKIVYELWAGSTDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKAASGNGLAEEKVALERLELQEERRAENARRIQRNPEI
VVDLITREKSVFDERDIAKVLYRYIDDAALFQNLMARLLQSPQTLRLDRERMDLVTGVRAPAKYTTRELI
RLEAQMANQAIWLSQRSSHRVNRTVLSGIVSRHDRLSDEQKTAIEHVAGPERIAAVIGRAGAGKTTMMKA
AREAWEAAGYRVVGGALAGKAADGLEKEAGIASRTLSAWELRWDQERDRLDEKSVFVLDEAGMVSSRQMA
RFVEAVTLSGAKLVLVGDPEQLQPIEAGAAFRAIAERIGYAELETIYRQREQWMRDASLDLARGNVSAAL
DAYAQRDLVRTGWTRDEAITALIAEWDHEYDPAKSTLILAHRRVDVRLLNEMARSKLVERGLIEAGHAFK
TEDGTRQFAAGDQIVFLKNEGSLGVKNGMLALVVDAQPGRIVAEIGNGEDRRRVVVEQRFYANVDHGYAT
TVHKSQGATVDRVKVLASSTLDRHLSYVAMTRHRETAELYVGLEEFAQRRGGVLIAHGEAPYEHKPGNRD
SYYVTLGFANGQERTVWGVDLARAMDASDSRIGDRIGLKHVGSQRVTLPNGTEVDRNSWKVVPIQELAMA
RLHERLSRAGSKETTLDYQDASHYRAALRFAEARGLHLMNVARTIAHDQLQWTVRQSSKLAELGARLVAV
AAKLGLGGAKSTVSTISVIKEAKPMVFGTTTFPRSIGQAVEDKLSADPGLKASWQEVSARFHHVFADPQA
AFKAVNVDAMLANGTVAATTIVRIAEQPESFGALKGKTGLFAGSAEKQARDTALVNAPALARDLQGFIAK
RAAAARRYEDEERAVRAQLSLDIPALSASGKQVLERVRDAIDRNDIPAGLEFALADKMVKAELEGFAKAV
SERFGERTFLPLAAKTADGKAFEVASAGMQPAQKNELRSAWDTIRTVQQLAAHERTAVALKQAEAIRQTQ
TKGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5937 | GenBank | WP_027690606 |
| Name | traG_QA638_RS39055_pCC283b_5 |
UniProt ID | _ |
| Length | 650 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 650 a.a. Molecular weight: 70599.44 Da Isoelectric Point: 9.7031
>WP_027690606.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTPKRILVAGIPAVAMLAIALTFPGIERWLSAFGTTDQAKLMLGRIGLALPYALAGACGVMFLFGTKGSI
NVKTSGWSVAAGGLGVIAVAALREGSRLLTFASQVPARRTLLSYADPSTIIGAGTALLVTFFALRVARMG
NAAFTRSEPRRIRGKRALHGEADWMTMQEAEKLFPETGGIVIGERYRVDRDDPAATSFRPGEPTSWGRGG
SAPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLVVLDPSNEVAPMVMEHRRKAGRRVIVLD
PKNAQSGFNALDWIGRHGGTKEEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDE
KHQTLRQVRANLSEPEPKLRARLQEIYDNSASDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYANYG
ALVSGSSFSTEALADGETDVFINIDLKALETHAGLARVIIGSFLNAIYHRDGAIKGRALFLLDEVARLGY
MRVLETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGTTT
VEIDQISRSFQSRGSSRTRSKQLASRQLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRSDMKSCVG
ENRFQQRGDRPEVSASAEPN
MTPKRILVAGIPAVAMLAIALTFPGIERWLSAFGTTDQAKLMLGRIGLALPYALAGACGVMFLFGTKGSI
NVKTSGWSVAAGGLGVIAVAALREGSRLLTFASQVPARRTLLSYADPSTIIGAGTALLVTFFALRVARMG
NAAFTRSEPRRIRGKRALHGEADWMTMQEAEKLFPETGGIVIGERYRVDRDDPAATSFRPGEPTSWGRGG
SAPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLVVLDPSNEVAPMVMEHRRKAGRRVIVLD
PKNAQSGFNALDWIGRHGGTKEEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDE
KHQTLRQVRANLSEPEPKLRARLQEIYDNSASDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYANYG
ALVSGSSFSTEALADGETDVFINIDLKALETHAGLARVIIGSFLNAIYHRDGAIKGRALFLLDEVARLGY
MRVLETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGTTT
VEIDQISRSFQSRGSSRTRSKQLASRQLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRSDMKSCVG
ENRFQQRGDRPEVSASAEPN
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5938 | GenBank | WP_027690590 |
| Name | t4cp2_QA638_RS39155_pCC283b_5 |
UniProt ID | _ |
| Length | 811 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 811 a.a. Molecular weight: 90951.41 Da Isoelectric Point: 5.8208
>WP_027690590.1 conjugal transfer protein TrbE [Rhizobium leguminosarum]
MVALRTFRSTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSESATDFERNELSRQINSILSRLG
TGWMIQVEAVRVPTTEYPSAERSHFPDAVTLLIDHERRRHFGQERGHFESRHALILTYRPPERRRSGLTR
YIYSDNESRSAKYADTALDAFRRSIREIEQYLGNVLSVQRMQTREVSERDGARVARYDELFQFIRFAITG
ENHPVRLPEIPMYLDWLATAELEHGLTPRVEGRFLGVIAIDGLPAESWPGILNSLNLMPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDESRSALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLVPLNSVWS
GQPFAPCPFYPPDAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFCRYEKAQVFA
FDKGNSMLTLTLGVGGDHYEIGGESGEGPSLAFCPLSELSTDADRAWASEWIETLVSLQGVTIAPDHRNA
ISRQIGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLTLGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNARQIEIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKADLARIRALSSTHGLEWPGQWLIERGINRNE
MVALRTFRSTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSESATDFERNELSRQINSILSRLG
TGWMIQVEAVRVPTTEYPSAERSHFPDAVTLLIDHERRRHFGQERGHFESRHALILTYRPPERRRSGLTR
YIYSDNESRSAKYADTALDAFRRSIREIEQYLGNVLSVQRMQTREVSERDGARVARYDELFQFIRFAITG
ENHPVRLPEIPMYLDWLATAELEHGLTPRVEGRFLGVIAIDGLPAESWPGILNSLNLMPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDESRSALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLVPLNSVWS
GQPFAPCPFYPPDAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFCRYEKAQVFA
FDKGNSMLTLTLGVGGDHYEIGGESGEGPSLAFCPLSELSTDADRAWASEWIETLVSLQGVTIAPDHRNA
ISRQIGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLTLGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNARQIEIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKADLARIRALSSTHGLEWPGQWLIERGINRNE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 112789..128018
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QA638_RS39075 (QA638_39075) | 111054..111617 | + | 564 | WP_050595854 | conjugative transfer signal peptidase TraF | - |
| QA638_RS39080 (QA638_39080) | 111607..112773 | + | 1167 | WP_027690603 | conjugal transfer protein TraB | - |
| QA638_RS39085 (QA638_39085) | 112789..113406 | + | 618 | WP_027690602 | TraH family protein | virB1 |
| QA638_RS39090 (QA638_39090) | 113430..114047 | - | 618 | WP_027690601 | hypothetical protein | - |
| QA638_RS39095 (QA638_39095) | 114044..114223 | - | 180 | WP_018010295 | hypothetical protein | - |
| QA638_RS39100 (QA638_39100) | 114373..115368 | - | 996 | WP_027690600 | alpha/beta hydrolase | - |
| QA638_RS39105 (QA638_39105) | 115358..116974 | - | 1617 | WP_027690599 | hypothetical protein | - |
| QA638_RS39110 (QA638_39110) | 117257..117961 | + | 705 | WP_027690598 | autoinducer binding domain-containing protein | - |
| QA638_RS39115 (QA638_39115) | 117962..118261 | - | 300 | WP_027690597 | transcriptional repressor TraM | - |
| QA638_RS39120 (QA638_39120) | 118462..119763 | - | 1302 | WP_027690596 | IncP-type conjugal transfer protein TrbI | virB10 |
| QA638_RS39125 (QA638_39125) | 119779..120225 | - | 447 | WP_027690595 | conjugal transfer protein TrbH | - |
| QA638_RS39130 (QA638_39130) | 120229..121059 | - | 831 | WP_027690594 | P-type conjugative transfer protein TrbG | virB9 |
| QA638_RS39135 (QA638_39135) | 121075..121737 | - | 663 | WP_027690593 | conjugal transfer protein TrbF | virB8 |
| QA638_RS39140 (QA638_39140) | 121759..122940 | - | 1182 | WP_027690592 | P-type conjugative transfer protein TrbL | virB6 |
| QA638_RS39145 (QA638_39145) | 122934..123134 | - | 201 | WP_027690591 | entry exclusion protein TrbK | - |
| QA638_RS39150 (QA638_39150) | 123131..123940 | - | 810 | WP_011654759 | P-type conjugative transfer protein TrbJ | virB5 |
| QA638_RS39155 (QA638_39155) | 123933..126368 | - | 2436 | WP_027690590 | conjugal transfer protein TrbE | virb4 |
| QA638_RS39160 (QA638_39160) | 126379..126678 | - | 300 | WP_011654761 | conjugal transfer protein TrbD | virB3 |
| QA638_RS39165 (QA638_39165) | 126671..127063 | - | 393 | WP_011654762 | TrbC/VirB2 family protein | virB2 |
| QA638_RS39170 (QA638_39170) | 127053..128018 | - | 966 | WP_027690589 | P-type conjugative transfer ATPase TrbB | virB11 |
Host bacterium
| ID | 8762 | GenBank | NZ_CP121640 |
| Plasmid name | pCC283b_5 | Incompatibility group | - |
| Plasmid size | 128397 bp | Coordinate of oriT [Strand] | 107343..107392 [+] |
| Host baterium | Rhizobium leguminosarum strain CC283b |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |