Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 108247 |
| Name | oriT_p1_EfsC94_repUS11 |
| Organism | Enterococcus faecalis strain EfsC94 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP124902 (9273..9322 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | 34..35 |
| Conserved sequence flanking the nic site |
GCTTGCAAAA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_p1_EfsC94_repUS11
AATATCGCAACATGCTAGCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
AATATCGCAACATGCTAGCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file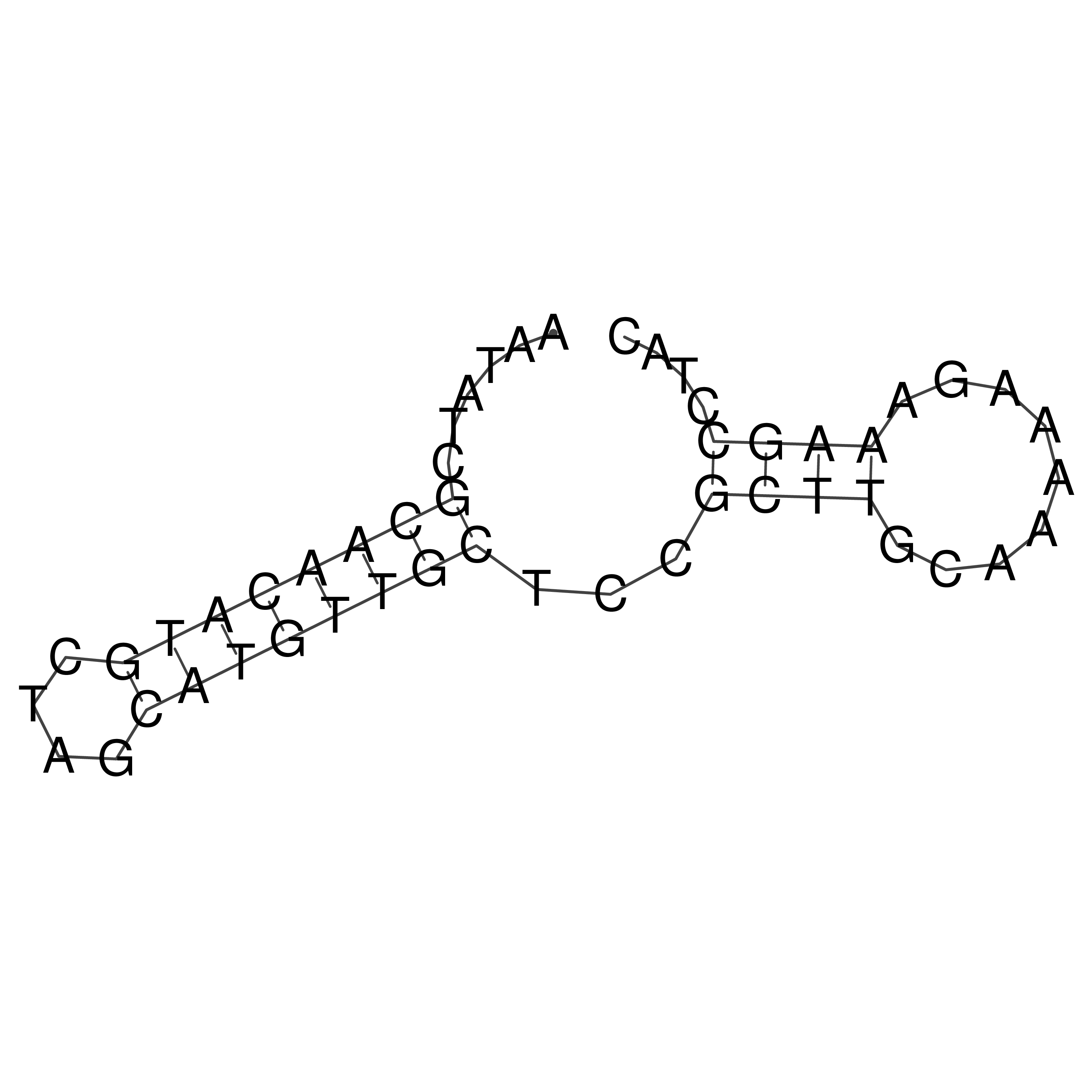
Relaxase
| ID | 5516 | GenBank | WP_161323729 |
| Name | Relaxase_QLQ35_RS13905_p1_EfsC94_repUS11 |
UniProt ID | _ |
| Length | 561 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65321.94 Da Isoelectric Point: 6.5183
>WP_161323729.1 relaxase/mobilization nuclease domain-containing protein [Enterococcus faecalis]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGKYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKRVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLDNGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGKYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKRVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLDNGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 2633 | GenBank | WP_002393737 |
| Name | WP_002393737_p1_EfsC94_repUS11 |
UniProt ID | A0A8E7CBI6 |
| Length | 118 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 118 a.a. Molecular weight: 13965.01 Da Isoelectric Point: 9.6545
>WP_002393737.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Enterococcus]
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKDQS
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKDQS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5884 | GenBank | WP_282861521 |
| Name | t4cp2_QLQ35_RS13885_p1_EfsC94_repUS11 |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 609 a.a. Molecular weight: 70193.23 Da Isoelectric Point: 6.2947
>WP_282861521.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecalis]
MQTYKKELKPYLYLGVFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGVLGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNYYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
MQTYKKELKPYLYLGVFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGVLGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNYYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 8682 | GenBank | NZ_CP124902 |
| Plasmid name | p1_EfsC94_repUS11 | Incompatibility group | - |
| Plasmid size | 78787 bp | Coordinate of oriT [Strand] | 9273..9322 [+] |
| Host baterium | Enterococcus faecalis strain EfsC94 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | prgB/asc10 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |