Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107973 |
| Name | oriT_2019-04-29292-1-3|4 |
| Organism | Providencia alcalifaciens strain 2019-04-29292-1-3 isolate 2019-04-29292-1-3 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_OU659165 (17632..17666 [-], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 9..15, 19..25 (GCAAAAT..ATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
>oriT_2019-04-29292-1-3|4
AAATTCTTGCAAAATGAAATTTTGCGTAGTGTGTG
AAATTCTTGCAAAATGAAATTTTGCGTAGTGTGTG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file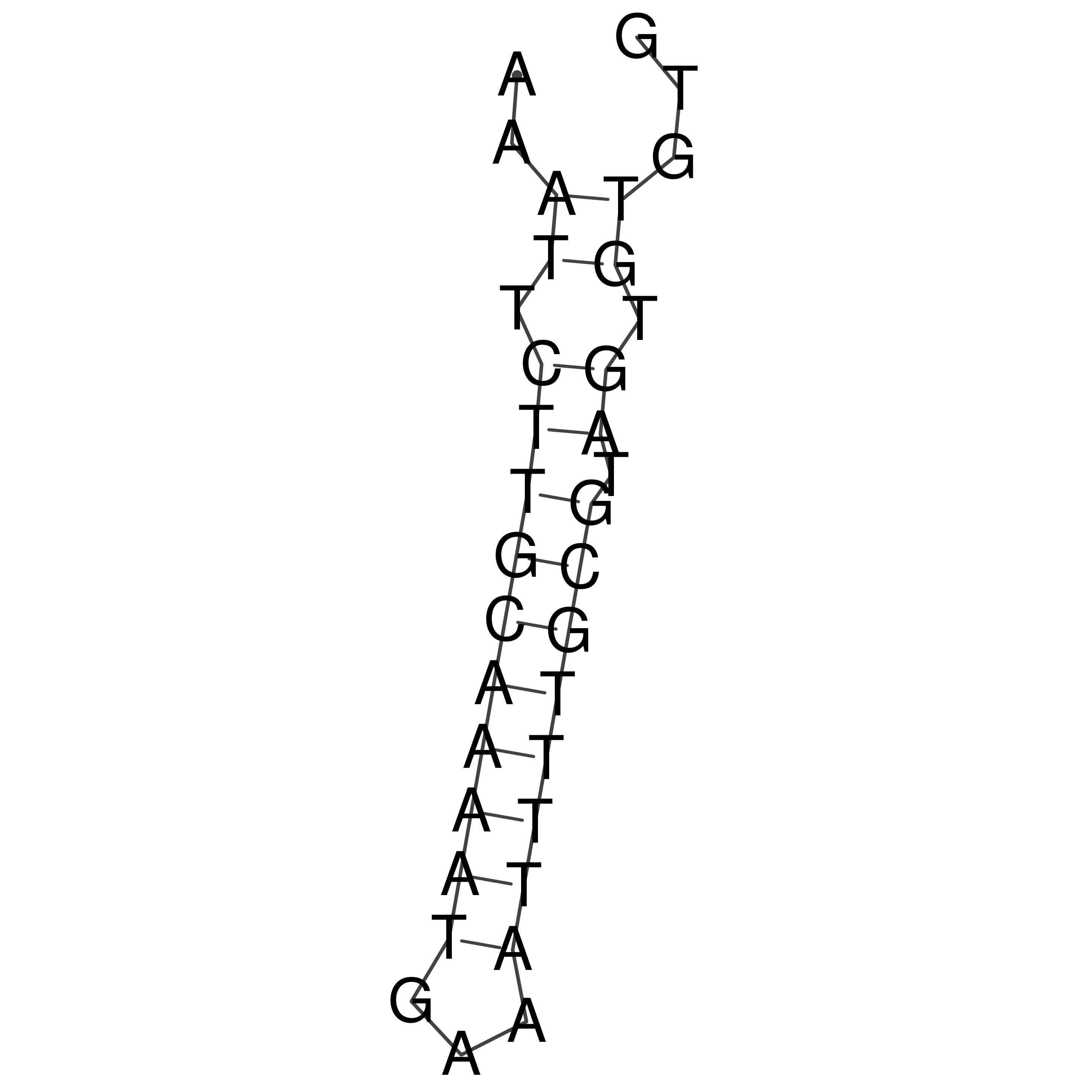
Relaxase
| ID | 5348 | GenBank | WP_224051218 |
| Name | traI_LDO74_RS20790_2019-04-29292-1-3|4 |
UniProt ID | _ |
| Length | 1708 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1708 a.a. Molecular weight: 187773.80 Da Isoelectric Point: 5.7090
>WP_224051218.1 conjugative transfer relaxase/helicase TraI [Providencia alcalifaciens]
MLTVAPIANNAANYYTHEDNYYFLGNLEARWMGEGAKSLGLEGEVTKDQLGEILEGRLPNDQSLERMENG
KNVHREGHDLTFSAPKSVSVLGIVLGDKRMIEAHNRAVEVALHEVEGLASTRMMVEGKTSLEMTKNLVVA
AFNHDTSREHDPQLHTHALVMNMTEHNGEWRTLSSDTRSKMGFSEALYDLRIALGEIYQNTLRKEIEGMG
FKTHNAGRNSLWEIDGVPVTPFSQRRQQILDAVGHEASLKSRDVAALDTRQAKTTPEKSELLTDWFDRLE
KNSFGADARKSFYAEAEQRLAKGQMGDASRDIQPDITRAVTDAIALLSDKQLSMTYSQVLAKTLGGLEAR
AGMIAQVREAIDVAIEQRQLIPLDEKKGLFTSSIHLMDELKLQQIATDMKQHNRTVSFQARTEALSPVME
KVADTLPSIAIVASQGLGKAQRDGVLDGITMAQSQGRDVAVMTLDTTSRQAFANDKRFAYVPKMTLGDGQ
PLKPNSTLVVANAEKLSLKSALSLLESAQSANVQVLMMDSGGRKGTGNVLATLQDAGVPRFQGESSAVLT
VKTAHIPDKNHRYAALAAEYAQLHSDMKSVVAQVSGVREQKIVTDTIRQTLTERGMLSEKTVTVTQLKPV
WLDSHSRKQIDSYREGMVMERWNADKKEVERYTIDRVTPETRSVTLLDSQGQKQVMKPQQFDGQWSAYQR
QTMDIAQGEQLTILAKQNKLSARDAVTVTGFMPNAIMVNFNGKTHRIDVRDGVKADYGYVTVPGQQANDT
GTVLLAASARDTQPTLLNTVARSGEHITVYTPLDKMETERRLSRSPVYQQARQLAGVEGLDSKAIEQAAD
KATEALLSKPEKALQQGMALAQESQVFFSRIDAIAKALPLHNSLDSQSLGKAFDRLVAQKAIIPVTSGKG
AAQQHYVTASTWAMEKQILTTILAGKDTQAPLMASVPEAQLNGLTSGQKAATELILGSRDQFVGIQGYAG
VGKTTQFTAVLKALDTLPATQRPEVIGLAPTHRAVGEMAAVGVKAQTLQAFLMDANQRQQLGETLDFKNT
LFLVDESSMIGNRNLSEVLQIIAQGGGRGVLSGDTAQLLSVDSGTPFALAQDRSALDTAIMKEIVRQNPE
LRPAIEAIIAGQVQMAMDTMNSVTPDVVPRRAGAELPEQSVIDAGDRVIARILDDYRGRTPEAQKETLIV
VQTNVDKDVINSGIHDILIEQGQLQGGKFVPILVRENTRTEALLSTAGMAEHAGKIALIQEQYYRIEVTQ
EGVSDGVVTLVDEQGKGHLLSAFESSLRDIGIYRQEARHIAAGEKINFTRTDKERGRVMNSDWTVSEVNE
HGQITLTNGEESRVLDPNSELTDQHLDYGYAGTAYKAQGASSPYVIVLGGVELGRRMLATLRDAYVALSR
TKAHVQTYSDDLDKWTSKITNPGNRKTAHDVLLAEQDYAAKVGNQLFDHAKPLNDTAIGRALSRQMGLGE
SHEGKFVYPSTKHPEPHVAWPAYDVHGKAQGTVLQAIELEGDKLQGLRTEGRLLGSEQAQFIVVKPSQNG
HTVIVNSLESAFETMAQQPEQGVVVQLNPDERLHTTMIEKITQGEVDNAYANPNTAQTDKADSDPKNLQT
PEEQAIDNALKEAEAALRQQANGEPKVPELSEDELRQVMAQERDSLLKGEHDLMEAKERVIEKAVQFERE
HQQQQRDVLRQQEREMVMEKTRDREFGD
MLTVAPIANNAANYYTHEDNYYFLGNLEARWMGEGAKSLGLEGEVTKDQLGEILEGRLPNDQSLERMENG
KNVHREGHDLTFSAPKSVSVLGIVLGDKRMIEAHNRAVEVALHEVEGLASTRMMVEGKTSLEMTKNLVVA
AFNHDTSREHDPQLHTHALVMNMTEHNGEWRTLSSDTRSKMGFSEALYDLRIALGEIYQNTLRKEIEGMG
FKTHNAGRNSLWEIDGVPVTPFSQRRQQILDAVGHEASLKSRDVAALDTRQAKTTPEKSELLTDWFDRLE
KNSFGADARKSFYAEAEQRLAKGQMGDASRDIQPDITRAVTDAIALLSDKQLSMTYSQVLAKTLGGLEAR
AGMIAQVREAIDVAIEQRQLIPLDEKKGLFTSSIHLMDELKLQQIATDMKQHNRTVSFQARTEALSPVME
KVADTLPSIAIVASQGLGKAQRDGVLDGITMAQSQGRDVAVMTLDTTSRQAFANDKRFAYVPKMTLGDGQ
PLKPNSTLVVANAEKLSLKSALSLLESAQSANVQVLMMDSGGRKGTGNVLATLQDAGVPRFQGESSAVLT
VKTAHIPDKNHRYAALAAEYAQLHSDMKSVVAQVSGVREQKIVTDTIRQTLTERGMLSEKTVTVTQLKPV
WLDSHSRKQIDSYREGMVMERWNADKKEVERYTIDRVTPETRSVTLLDSQGQKQVMKPQQFDGQWSAYQR
QTMDIAQGEQLTILAKQNKLSARDAVTVTGFMPNAIMVNFNGKTHRIDVRDGVKADYGYVTVPGQQANDT
GTVLLAASARDTQPTLLNTVARSGEHITVYTPLDKMETERRLSRSPVYQQARQLAGVEGLDSKAIEQAAD
KATEALLSKPEKALQQGMALAQESQVFFSRIDAIAKALPLHNSLDSQSLGKAFDRLVAQKAIIPVTSGKG
AAQQHYVTASTWAMEKQILTTILAGKDTQAPLMASVPEAQLNGLTSGQKAATELILGSRDQFVGIQGYAG
VGKTTQFTAVLKALDTLPATQRPEVIGLAPTHRAVGEMAAVGVKAQTLQAFLMDANQRQQLGETLDFKNT
LFLVDESSMIGNRNLSEVLQIIAQGGGRGVLSGDTAQLLSVDSGTPFALAQDRSALDTAIMKEIVRQNPE
LRPAIEAIIAGQVQMAMDTMNSVTPDVVPRRAGAELPEQSVIDAGDRVIARILDDYRGRTPEAQKETLIV
VQTNVDKDVINSGIHDILIEQGQLQGGKFVPILVRENTRTEALLSTAGMAEHAGKIALIQEQYYRIEVTQ
EGVSDGVVTLVDEQGKGHLLSAFESSLRDIGIYRQEARHIAAGEKINFTRTDKERGRVMNSDWTVSEVNE
HGQITLTNGEESRVLDPNSELTDQHLDYGYAGTAYKAQGASSPYVIVLGGVELGRRMLATLRDAYVALSR
TKAHVQTYSDDLDKWTSKITNPGNRKTAHDVLLAEQDYAAKVGNQLFDHAKPLNDTAIGRALSRQMGLGE
SHEGKFVYPSTKHPEPHVAWPAYDVHGKAQGTVLQAIELEGDKLQGLRTEGRLLGSEQAQFIVVKPSQNG
HTVIVNSLESAFETMAQQPEQGVVVQLNPDERLHTTMIEKITQGEVDNAYANPNTAQTDKADSDPKNLQT
PEEQAIDNALKEAEAALRQQANGEPKVPELSEDELRQVMAQERDSLLKGEHDLMEAKERVIEKAVQFERE
HQQQQRDVLRQQEREMVMEKTRDREFGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5633 | GenBank | WP_224051197 |
| Name | traC_LDO74_RS20695_2019-04-29292-1-3|4 |
UniProt ID | _ |
| Length | 903 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 903 a.a. Molecular weight: 101474.94 Da Isoelectric Point: 6.5159
>WP_224051197.1 type IV secretion system protein TraC [Providencia alcalifaciens]
MNEKPSLIDKLLAPFLPKSKDIVFDDEPEPVSLQEALFKNLFTAWREPDGNKRAAEALENMDYPSFCSLL
PYRNYDAKSGLFINKTTIGFALECTPLVGANEKIVDALDYFLRNKLPRKIPLTFMLLGSKCISEQLGYGL
KDFGWQGEEAEKFNHITRAFYERGALVGLDNKLNLPLSLRNYRLFVCYAEEHKKADALTLQKVNQILSIV
MKSLAGASLHCEQMNDKGLLQLVREISNFRHDQIALPSSQVDPYESLNDQCADRSLTLEIRPDAIHQSLS
MGKGQKSQTRIMNYMLEKNPTQFALWQTGDNLSNLLDPASSVSCPFAITMTVYVEDLVKTQNEAKKKFIT
NNSRANSKYINWVPGVKRKAEEWGALREGLDTNQMALARYSYGITLFCEDDDDKALEAEMALMNTYTTNH
LKLCPPTFMQLRNYLALFPFVMQEGLWGDMVRSGGTLRAKAFNVANLLPVVADNRICQKGLPIPSYRHQL
SFLDIFERGSGLGNDNFNAAVCGTSGSGKSFLMQALIRQVLDAGGWAYVLDMGDSYKELCHSLGGVYVDA
RDLKFNPFAGVVDIKETAESIRDLLIVLANPSGDMDDVSKSILLNAVQEVWEGKGPNGRKGNKVLIDDVH
DYLEAEVKAAYFDPQSTVQNKMQEIIVALKKYTTKGLYGEYFNSDKPALNEQVRFTVLEMGGLQNKPDLL
AAVMFSMMIFIQQRMYLTARSIKKLAAIDEGWKLLDNKSRFVADFIESGYRTARKYGGSYITISQGIDDF
DGDGASTAAKAAWSNSSFKIILRQNMEAFRKYNQKNPDQFNPVERAIIEGFPAAGDAHFSAFMLRIAGRS
SFHRLVTDPISRALYSTDGDDFQFRENLMAEGLSQQAALLALAKRKFPQEMETLSQWQSQTRH
MNEKPSLIDKLLAPFLPKSKDIVFDDEPEPVSLQEALFKNLFTAWREPDGNKRAAEALENMDYPSFCSLL
PYRNYDAKSGLFINKTTIGFALECTPLVGANEKIVDALDYFLRNKLPRKIPLTFMLLGSKCISEQLGYGL
KDFGWQGEEAEKFNHITRAFYERGALVGLDNKLNLPLSLRNYRLFVCYAEEHKKADALTLQKVNQILSIV
MKSLAGASLHCEQMNDKGLLQLVREISNFRHDQIALPSSQVDPYESLNDQCADRSLTLEIRPDAIHQSLS
MGKGQKSQTRIMNYMLEKNPTQFALWQTGDNLSNLLDPASSVSCPFAITMTVYVEDLVKTQNEAKKKFIT
NNSRANSKYINWVPGVKRKAEEWGALREGLDTNQMALARYSYGITLFCEDDDDKALEAEMALMNTYTTNH
LKLCPPTFMQLRNYLALFPFVMQEGLWGDMVRSGGTLRAKAFNVANLLPVVADNRICQKGLPIPSYRHQL
SFLDIFERGSGLGNDNFNAAVCGTSGSGKSFLMQALIRQVLDAGGWAYVLDMGDSYKELCHSLGGVYVDA
RDLKFNPFAGVVDIKETAESIRDLLIVLANPSGDMDDVSKSILLNAVQEVWEGKGPNGRKGNKVLIDDVH
DYLEAEVKAAYFDPQSTVQNKMQEIIVALKKYTTKGLYGEYFNSDKPALNEQVRFTVLEMGGLQNKPDLL
AAVMFSMMIFIQQRMYLTARSIKKLAAIDEGWKLLDNKSRFVADFIESGYRTARKYGGSYITISQGIDDF
DGDGASTAAKAAWSNSSFKIILRQNMEAFRKYNQKNPDQFNPVERAIIEGFPAAGDAHFSAFMLRIAGRS
SFHRLVTDPISRALYSTDGDDFQFRENLMAEGLSQQAALLALAKRKFPQEMETLSQWQSQTRH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5634 | GenBank | WP_282563892 |
| Name | traD_LDO74_RS20785_2019-04-29292-1-3|4 |
UniProt ID | _ |
| Length | 742 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 742 a.a. Molecular weight: 84918.85 Da Isoelectric Point: 5.1648
>WP_282563892.1 type IV conjugative transfer system coupling protein TraD [Providencia alcalifaciens]
MSSTKHITQGGQVFSYMLGMFMQVNKRISFWLILLFLMIFPIIFYFKVPLQEIKNGGLYWWLYLTAGGEK
ALYRVAPIYEVDFDGQLYRFTSDAILKDDYMIYAGNIVLQELGLSLLWACIIVIVLFFVIYRVFHGLGKR
QANDDVIGGRTLTEDIKHVRNMLHARREASPITFDGLPLKLNSEVQNMLMHGTPGSGKSNTINKLLIQLR
KRGDMVIVFDKGCSLVKRHYNENIDKLLNPLDERCENWDLWGECLTTPDFDSVANTLIPQSTSEDPFWTG
SARTIFTTVAAKLGSDDQRSYNKLLRTLLAIDLKSLRAYVEKTEASNLMEEKVEKTAISIRGVLTNYVKA
LRYLQGIERNGKAPFAIREWMKSVNEPKQRQGWLWITSNARQHESLKPLISMWLAQAANCLLEMGENPER
RVWFIYDELASLHKLPELPQVLSEARKFGGCFILGFQNKPQLDYTYGHDYANAMMDLLNTRFFFRSPDES
VAKWVRDQLGQTRKKVASEQYSFGPETVRDGVSFSKQQEDVDLVNYSDVQSLPDLDCYITLPGQYPVVKI
SMRYEKIKPIAAEFTPRPINDSLDQEIEAEIKSRMQDDALPAVMNQIIDNLISGKPIETPVDTAQNNTAL
EVTQTQPQSTVELTSQSVTQNSTPITTETAASDNPPENQGGQEVIIDNVKVNTETGEILGRVIDEEQNPQ
DLYDSYTRMRQDEKEIFNRPKPSQEKPKHINDDHDEPEMWWC
MSSTKHITQGGQVFSYMLGMFMQVNKRISFWLILLFLMIFPIIFYFKVPLQEIKNGGLYWWLYLTAGGEK
ALYRVAPIYEVDFDGQLYRFTSDAILKDDYMIYAGNIVLQELGLSLLWACIIVIVLFFVIYRVFHGLGKR
QANDDVIGGRTLTEDIKHVRNMLHARREASPITFDGLPLKLNSEVQNMLMHGTPGSGKSNTINKLLIQLR
KRGDMVIVFDKGCSLVKRHYNENIDKLLNPLDERCENWDLWGECLTTPDFDSVANTLIPQSTSEDPFWTG
SARTIFTTVAAKLGSDDQRSYNKLLRTLLAIDLKSLRAYVEKTEASNLMEEKVEKTAISIRGVLTNYVKA
LRYLQGIERNGKAPFAIREWMKSVNEPKQRQGWLWITSNARQHESLKPLISMWLAQAANCLLEMGENPER
RVWFIYDELASLHKLPELPQVLSEARKFGGCFILGFQNKPQLDYTYGHDYANAMMDLLNTRFFFRSPDES
VAKWVRDQLGQTRKKVASEQYSFGPETVRDGVSFSKQQEDVDLVNYSDVQSLPDLDCYITLPGQYPVVKI
SMRYEKIKPIAAEFTPRPINDSLDQEIEAEIKSRMQDDALPAVMNQIIDNLISGKPIETPVDTAQNNTAL
EVTQTQPQSTVELTSQSVTQNSTPITTETAASDNPPENQGGQEVIIDNVKVNTETGEILGRVIDEEQNPQ
DLYDSYTRMRQDEKEIFNRPKPSQEKPKHINDDHDEPEMWWC
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 17049..47612
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LDO74_RS20575 (NVI2019_OGMBKCAO_04136) | 13211..13501 | + | 291 | WP_224051205 | hypothetical protein | - |
| LDO74_RS20580 (NVI2019_OGMBKCAO_04137) | 13565..15565 | + | 2001 | WP_224051204 | ParB/RepB/Spo0J family partition protein | - |
| LDO74_RS20585 (NVI2019_OGMBKCAO_04138) | 15607..16290 | + | 684 | WP_224051203 | plasmid SOS inhibition protein A | - |
| LDO74_RS20590 (NVI2019_OGMBKCAO_04140) | 16512..16943 | + | 432 | WP_263422653 | LPD29 domain-containing protein | - |
| LDO74_RS20595 (NVI2019_OGMBKCAO_04141) | 17049..17501 | - | 453 | WP_224051202 | transglycosylase SLT domain-containing protein | virB1 |
| LDO74_RS20600 (NVI2019_OGMBKCAO_04142) | 17976..18350 | + | 375 | WP_036965543 | relaxosome protein TraM | - |
| LDO74_RS20605 (NVI2019_OGMBKCAO_04143) | 18359..18676 | + | 318 | WP_036965545 | hypothetical protein | - |
| LDO74_RS20610 (NVI2019_OGMBKCAO_04144) | 18814..19533 | + | 720 | WP_036965547 | hypothetical protein | - |
| LDO74_RS20615 (NVI2019_OGMBKCAO_04145) | 19574..19765 | + | 192 | WP_036965549 | TraY domain-containing protein | - |
| LDO74_RS20620 (NVI2019_OGMBKCAO_04146) | 20039..20407 | + | 369 | WP_036965551 | type IV conjugative transfer system pilin TraA | - |
| LDO74_RS20625 (NVI2019_OGMBKCAO_04147) | 20426..20731 | + | 306 | WP_071548954 | type IV conjugative transfer system protein TraL | traL |
| LDO74_RS20630 (NVI2019_OGMBKCAO_04148) | 20745..21314 | + | 570 | WP_036965554 | type IV conjugative transfer system protein TraE | traE |
| LDO74_RS20635 (NVI2019_OGMBKCAO_04149) | 21298..22071 | + | 774 | WP_036965555 | type-F conjugative transfer system secretin TraK | traK |
| LDO74_RS20640 (NVI2019_OGMBKCAO_04150) | 22058..23419 | + | 1362 | WP_224051200 | TrbI/VirB10 family protein | traB |
| LDO74_RS21770 (NVI2019_OGMBKCAO_04151) | 23489..23683 | + | 195 | WP_036965558 | hypothetical protein | - |
| LDO74_RS20650 (NVI2019_OGMBKCAO_04152) | 23690..24181 | + | 492 | WP_036965560 | transcription/translation regulatory transformer protein RfaH | - |
| LDO74_RS20655 (NVI2019_OGMBKCAO_04153) | 24202..24540 | + | 339 | WP_036965562 | hypothetical protein | - |
| LDO74_RS20660 (NVI2019_OGMBKCAO_04154) | 24542..24898 | + | 357 | WP_036965564 | hypothetical protein | - |
| LDO74_RS20665 (NVI2019_OGMBKCAO_04155) | 24907..25182 | + | 276 | WP_036965565 | hypothetical protein | - |
| LDO74_RS20670 (NVI2019_OGMBKCAO_04156) | 25209..25505 | + | 297 | WP_036965567 | hypothetical protein | - |
| LDO74_RS20675 (NVI2019_OGMBKCAO_04157) | 25598..25999 | + | 402 | WP_036965569 | hypothetical protein | - |
| LDO74_RS21775 (NVI2019_OGMBKCAO_04158) | 26084..26401 | + | 318 | WP_224051199 | hypothetical protein | - |
| LDO74_RS20685 (NVI2019_OGMBKCAO_04159) | 26372..27250 | - | 879 | WP_070929355 | hypothetical protein | - |
| LDO74_RS20690 (NVI2019_OGMBKCAO_04160) | 27297..27860 | + | 564 | WP_224051198 | type IV conjugative transfer system lipoprotein TraV | traV |
| LDO74_RS20695 (NVI2019_OGMBKCAO_04161) | 27862..30573 | + | 2712 | WP_224051197 | type IV secretion system protein TraC | virb4 |
| LDO74_RS20700 (NVI2019_OGMBKCAO_04162) | 30549..30974 | + | 426 | WP_224051196 | type-F conjugative transfer system protein TrbI | - |
| LDO74_RS20705 (NVI2019_OGMBKCAO_04163) | 30984..31616 | + | 633 | WP_224051195 | type-F conjugative transfer system protein TraW | - |
| LDO74_RS20710 (NVI2019_OGMBKCAO_04164) | 31613..32608 | + | 996 | WP_036965577 | conjugal transfer pilus assembly protein TraU | traU |
| LDO74_RS20715 (NVI2019_OGMBKCAO_04165) | 32624..33241 | + | 618 | WP_224051194 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LDO74_RS20720 (NVI2019_OGMBKCAO_04166) | 33241..35085 | + | 1845 | WP_224051193 | conjugal transfer protein TraN | - |
| LDO74_RS20725 (NVI2019_OGMBKCAO_04167) | 35082..35870 | + | 789 | WP_224051192 | type-F conjugative transfer system pilin assembly protein TraF | - |
| LDO74_RS20730 (NVI2019_OGMBKCAO_04168) | 35885..36277 | + | 393 | WP_036965583 | hypothetical protein | - |
| LDO74_RS20735 (NVI2019_OGMBKCAO_04169) | 36265..37200 | - | 936 | WP_036965584 | hypothetical protein | - |
| LDO74_RS20740 (NVI2019_OGMBKCAO_04170) | 37359..37658 | + | 300 | WP_036965586 | hypothetical protein | - |
| LDO74_RS20745 (NVI2019_OGMBKCAO_04171) | 37664..38188 | + | 525 | WP_071992030 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LDO74_RS21780 (NVI2019_OGMBKCAO_04172) | 38200..38628 | + | 429 | WP_036965590 | hypothetical protein | - |
| LDO74_RS20755 (NVI2019_OGMBKCAO_04173) | 38625..39020 | + | 396 | WP_224051220 | hypothetical protein | - |
| LDO74_RS20760 (NVI2019_OGMBKCAO_04174) | 39217..40593 | + | 1377 | WP_036965592 | conjugal transfer pilus assembly protein TraH | traH |
| LDO74_RS20765 (NVI2019_OGMBKCAO_04175) | 40606..43494 | + | 2889 | WP_051473561 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LDO74_RS20770 (NVI2019_OGMBKCAO_04176) | 43673..44401 | + | 729 | WP_036965594 | complement resistance protein TraT | - |
| LDO74_RS20775 (NVI2019_OGMBKCAO_04177) | 44522..44728 | + | 207 | WP_036965596 | TraR/DksA C4-type zinc finger protein | - |
| LDO74_RS20780 (NVI2019_OGMBKCAO_04178) | 44783..45172 | + | 390 | WP_155251352 | hypothetical protein | - |
| LDO74_RS20785 (NVI2019_OGMBKCAO_04179) | 45384..47606 | + | 2223 | WP_224051219 | type IV conjugative transfer system coupling protein TraD | - |
Host bacterium
| ID | 8408 | GenBank | NZ_OU659165 |
| Plasmid name | 2019-04-29292-1-3|4 | Incompatibility group | - |
| Plasmid size | 57474 bp | Coordinate of oriT [Strand] | 17632..17666 [-] |
| Host baterium | Providencia alcalifaciens strain 2019-04-29292-1-3 isolate 2019-04-29292-1-3 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |