Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107947 |
| Name | oriT_FDAARGOS_633|unnamed1 |
| Organism | Agrobacterium pusense strain FDAARGOS_633 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP050897 (205805..205841 [-], 37 nt) |
| oriT length | 37 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 37 nt
>oriT_FDAARGOS_633|unnamed1
GGATCAAAGGGCGCAATTATACGTCGCTGCCGCGACG
GGATCAAAGGGCGCAATTATACGTCGCTGCCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file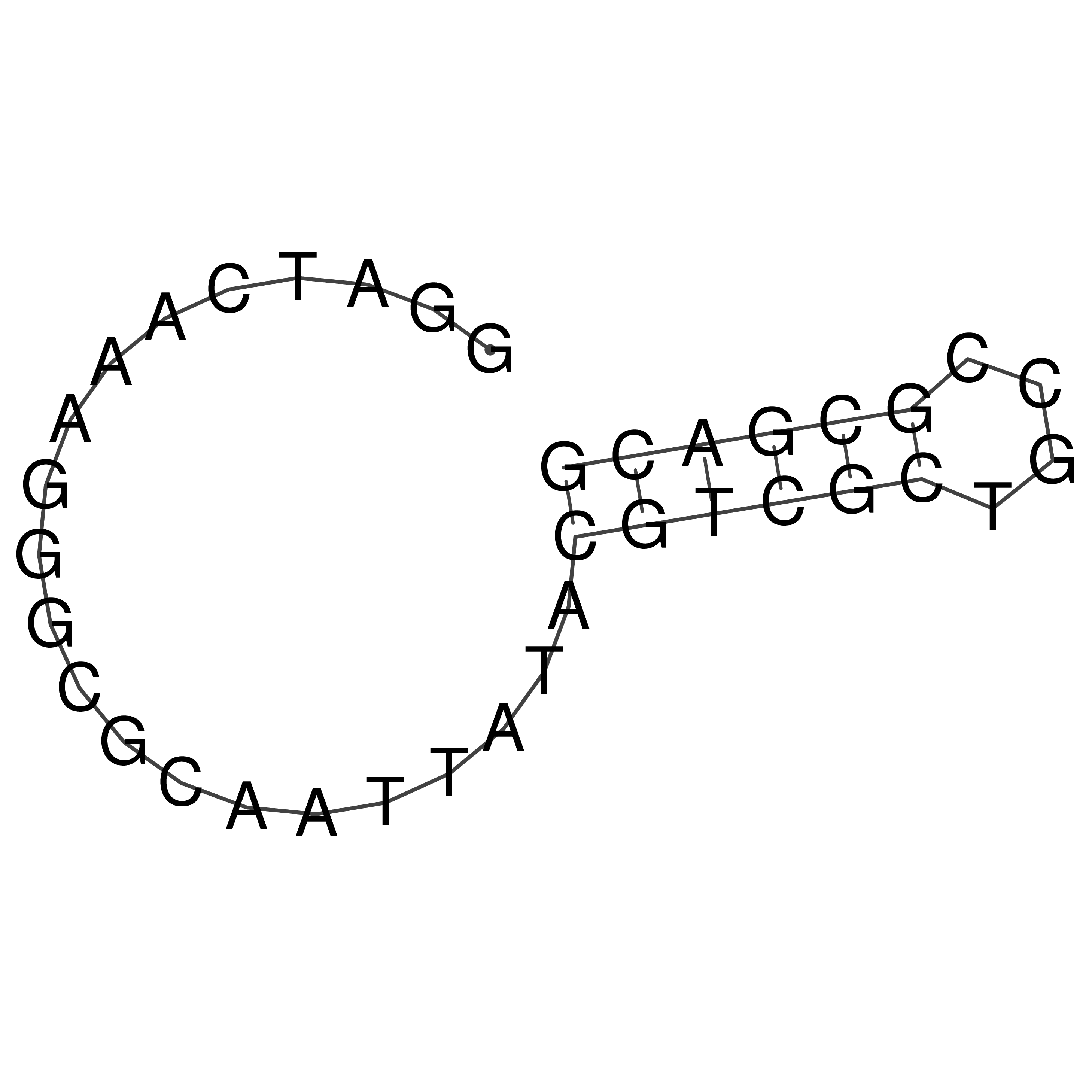
Relaxase
| ID | 5334 | GenBank | WP_080823885 |
| Name | traA_FOB41_RS02465_FDAARGOS_633|unnamed1 |
UniProt ID | _ |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 121552.03 Da Isoelectric Point: 10.1056
>WP_080823885.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Hyphomicrobiales]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKLGLIHEEFVIPEDAPAWVRAMIADR
SISGASEVFWNKVEAFEKRADAQLAKDVTIALPIELTADQNIALVRDFVERHVTAKGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGAKKVAVIGPDGKPVRNDAGKIVYELWAGSADDFNVFRDGWFACHNRHLALAG
MDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKAQDAKIGLALERLELQEERRAQNARRIQRHPEIVLDL
VTREKSVFDERDIAKILYRYIDDVALFQTLMVRILQSPQTLRLDRERMDLVTGVKAPAKYTTRELIRIEA
EMANRAIWLSQRSSHRVRDLVLKSVFARHERLSDEQRTAIEHVAGHERIAAVIGRAGAGKTTMMKAAREA
WESAGYRVVGGALAGKAAEGLSSEAGIASRTLSSWQLRWSQGRSQLDDRTVFVLDEAGMVSSRQMALFVE
VATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRERWMRDASLDLARGRVGKAVDAYR
ANGKLMGSELKTDAVINLISDWSRDYDPTKSSLILAHLRRDIRMLNQLARGKLVDRGIIDIGHSFKTADG
NRNFAAGDQVVFLKNEGSLGVKNGMLAKVVDAAPGRLVVQIGGAGNPRLVTVEQRFYNNIDHGYATTVHK
SQGATVDHVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKAGGLVPILSRSNAKETTLDYAAGRFY
AQALRFAEARGLHLVNIARTIAHDQLRWTIRQSSRLAELGVRLAAVAAKLGLGTTKLDPSTSAAIKEGKP
MVSGTTIFPHSIGQAIEDKLSADPALKADWQEVSARFHHIYADPQAAFKAVNVDAMLVDGAAAKATIIKI
AEQPESFGSLKGKTGLFAGRADRQARDNALINAPALARDLESFISKRSEAATRYEAEERALRSKLSLDIP
ALSASAHQVLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKIVAERFGERAFLPLAAKTADGKAFKS
LSVGMQQAQKSELRSAWDTMRTVQQLAAHERSAVALKHAETLRQTQSKGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKLGLIHEEFVIPEDAPAWVRAMIADR
SISGASEVFWNKVEAFEKRADAQLAKDVTIALPIELTADQNIALVRDFVERHVTAKGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGAKKVAVIGPDGKPVRNDAGKIVYELWAGSADDFNVFRDGWFACHNRHLALAG
MDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKAQDAKIGLALERLELQEERRAQNARRIQRHPEIVLDL
VTREKSVFDERDIAKILYRYIDDVALFQTLMVRILQSPQTLRLDRERMDLVTGVKAPAKYTTRELIRIEA
EMANRAIWLSQRSSHRVRDLVLKSVFARHERLSDEQRTAIEHVAGHERIAAVIGRAGAGKTTMMKAAREA
WESAGYRVVGGALAGKAAEGLSSEAGIASRTLSSWQLRWSQGRSQLDDRTVFVLDEAGMVSSRQMALFVE
VATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRERWMRDASLDLARGRVGKAVDAYR
ANGKLMGSELKTDAVINLISDWSRDYDPTKSSLILAHLRRDIRMLNQLARGKLVDRGIIDIGHSFKTADG
NRNFAAGDQVVFLKNEGSLGVKNGMLAKVVDAAPGRLVVQIGGAGNPRLVTVEQRFYNNIDHGYATTVHK
SQGATVDHVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKAGGLVPILSRSNAKETTLDYAAGRFY
AQALRFAEARGLHLVNIARTIAHDQLRWTIRQSSRLAELGVRLAAVAAKLGLGTTKLDPSTSAAIKEGKP
MVSGTTIFPHSIGQAIEDKLSADPALKADWQEVSARFHHIYADPQAAFKAVNVDAMLVDGAAAKATIIKI
AEQPESFGSLKGKTGLFAGRADRQARDNALINAPALARDLESFISKRSEAATRYEAEERALRSKLSLDIP
ALSASAHQVLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKIVAERFGERAFLPLAAKTADGKAFKS
LSVGMQQAQKSELRSAWDTMRTVQQLAAHERSAVALKHAETLRQTQSKGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5605 | GenBank | WP_092938694 |
| Name | t4cp2_FOB41_RS02400_FDAARGOS_633|unnamed1 |
UniProt ID | _ |
| Length | 811 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 811 a.a. Molecular weight: 91187.65 Da Isoelectric Point: 5.8000
>WP_092938694.1 MULTISPECIES: conjugal transfer protein TrbE [Hyphomicrobiales]
MVALRSFRSAGPSFSELVPYAGIVDNGIILLKDGSLMAGWYFAGPDSESATDFERNELSRQINATLSRLG
SGWMIQVEAVRFPTIDYPSADRSHFPDAVTRMIDDERRNHFAREQGHFESRHALIMTYRPPERRRSGLTR
YIYSDTESRTASYADTVLETFRRSIREIEQYLGNSVSIQRMLTREVPERGGARVARYDDLFQFIRFTITG
ENHPIRLPEIPMYLDWLVTAEMEHGLTPRVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMAMVMETEDAIAEASSQLVAYGYYTPVVIL
FDERLEALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLIPLNSVWS
GSPTAPCPFYPHDAPPLMQVASGSTPFRLNLHVNDVGHSLIFGPTGSGKSTLLALIAAQFCRYEQAQVFA
FDKGNSMLPLTLAVEGDHYAIGAEDGEGAKLAFCPLSDLSSDGDRAWASEWIETLVALQGVTINPDHRNA
ISRQIGLMAAAPGRSLSDFVSGVQARDIKDALHHYTVDGPMGQLLDAEHDGLALGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNERQIDIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKSDLARIRALQSAHGKEWPGQWLIERGINRYA
MVALRSFRSAGPSFSELVPYAGIVDNGIILLKDGSLMAGWYFAGPDSESATDFERNELSRQINATLSRLG
SGWMIQVEAVRFPTIDYPSADRSHFPDAVTRMIDDERRNHFAREQGHFESRHALIMTYRPPERRRSGLTR
YIYSDTESRTASYADTVLETFRRSIREIEQYLGNSVSIQRMLTREVPERGGARVARYDDLFQFIRFTITG
ENHPIRLPEIPMYLDWLVTAEMEHGLTPRVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMAMVMETEDAIAEASSQLVAYGYYTPVVIL
FDERLEALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLIPLNSVWS
GSPTAPCPFYPHDAPPLMQVASGSTPFRLNLHVNDVGHSLIFGPTGSGKSTLLALIAAQFCRYEQAQVFA
FDKGNSMLPLTLAVEGDHYAIGAEDGEGAKLAFCPLSDLSSDGDRAWASEWIETLVALQGVTINPDHRNA
ISRQIGLMAAAPGRSLSDFVSGVQARDIKDALHHYTVDGPMGQLLDAEHDGLALGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNERQIDIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKSDLARIRALQSAHGKEWPGQWLIERGINRYA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5606 | GenBank | WP_080823883 |
| Name | traG_FOB41_RS02480_FDAARGOS_633|unnamed1 |
UniProt ID | _ |
| Length | 653 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 653 a.a. Molecular weight: 70612.43 Da Isoelectric Point: 9.3955
>WP_080823883.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Hyphomicrobiales]
MTPKKIFLAAAPAVAMLAVVFAVPGIEHWVAGFGTTSEGQVMLGRIGLALPYALAAACGVIFLFGARGSI
DIRTAGWSVATGGLGVVLVGAFREGARLLSFASQVPAGQSAMSYADPSTVLGGGLAILATFFALRVVRMG
NAAFARSEPKRIRGRRALHGEADWMSMQEAEKLLPETGGIVIGERYRVDRDSPAAVSFRAAEPATWGRGG
SAPLLCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGGSLVVLDPSNEVAPMVMDHRRKSGRRVIVLD
PKIAGSGFNALDWIGRHGGTREEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDE
KDQTLRQVRANLSEPEPKLRARLQEIYDNSASDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYTNFG
ALVSGSNFSTDALANGDTDVFINIDLKTLETHAGLARVVIGSFLNAIYNRNGAMKSRTLFLLDEAARLGY
MRVLETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGTTT
VEIDQVSRSFQSRGSSRTRSKQLASRPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRNDMKTCVM
QNPFHRPAALPHGVFDQRQADQE
MTPKKIFLAAAPAVAMLAVVFAVPGIEHWVAGFGTTSEGQVMLGRIGLALPYALAAACGVIFLFGARGSI
DIRTAGWSVATGGLGVVLVGAFREGARLLSFASQVPAGQSAMSYADPSTVLGGGLAILATFFALRVVRMG
NAAFARSEPKRIRGRRALHGEADWMSMQEAEKLLPETGGIVIGERYRVDRDSPAAVSFRAAEPATWGRGG
SAPLLCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGGSLVVLDPSNEVAPMVMDHRRKSGRRVIVLD
PKIAGSGFNALDWIGRHGGTREEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDE
KDQTLRQVRANLSEPEPKLRARLQEIYDNSASDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYTNFG
ALVSGSNFSTDALANGDTDVFINIDLKTLETHAGLARVVIGSFLNAIYNRNGAMKSRTLFLLDEAARLGY
MRVLETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGTTT
VEIDQVSRSFQSRGSSRTRSKQLASRPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRNDMKTCVM
QNPFHRPAALPHGVFDQRQADQE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 189161..200689
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FOB41_RS02370 (FOB41_02385) | 185124..186341 | - | 1218 | WP_080823900 | plasmid replication protein RepC | - |
| FOB41_RS02375 (FOB41_02390) | 186516..187475 | - | 960 | WP_080823899 | plasmid partitioning protein RepB | - |
| FOB41_RS02380 (FOB41_02395) | 187596..188792 | - | 1197 | WP_003501703 | plasmid partitioning protein RepA | - |
| FOB41_RS02385 (FOB41_02400) | 189161..190126 | + | 966 | WP_080823898 | P-type conjugative transfer ATPase TrbB | virB11 |
| FOB41_RS02390 (FOB41_02405) | 190116..190502 | + | 387 | WP_035243728 | TrbC/VirB2 family protein | virB2 |
| FOB41_RS02395 (FOB41_02410) | 190495..190794 | + | 300 | WP_080823897 | conjugal transfer protein TrbD | virB3 |
| FOB41_RS02400 (FOB41_02415) | 190804..193239 | + | 2436 | WP_092938694 | conjugal transfer protein TrbE | virb4 |
| FOB41_RS02405 (FOB41_02420) | 193232..194041 | + | 810 | WP_080823896 | P-type conjugative transfer protein TrbJ | virB5 |
| FOB41_RS02410 (FOB41_02425) | 194038..194241 | + | 204 | WP_080823895 | entry exclusion protein TrbK | - |
| FOB41_RS02415 (FOB41_02430) | 194235..195416 | + | 1182 | WP_080823894 | P-type conjugative transfer protein TrbL | virB6 |
| FOB41_RS02420 (FOB41_02435) | 195437..196099 | + | 663 | WP_003501718 | conjugal transfer protein TrbF | virB8 |
| FOB41_RS02425 (FOB41_02440) | 196115..196831 | + | 717 | Protein_181 | P-type conjugative transfer protein TrbG | - |
| FOB41_RS02430 (FOB41_02445) | 196834..197283 | + | 450 | Protein_182 | recombinase family protein | - |
| FOB41_RS02435 (FOB41_02450) | 197273..198124 | + | 852 | WP_080823890 | protein-L-isoaspartate O-methyltransferase | - |
| FOB41_RS02440 (FOB41_02455) | 198792..199091 | + | 300 | WP_080823889 | transcriptional repressor TraM | - |
| FOB41_RS02445 (FOB41_02460) | 199102..199806 | - | 705 | WP_003501732 | transcriptional regulator TraR | - |
| FOB41_RS02450 (FOB41_02465) | 200075..200689 | - | 615 | WP_080823888 | TraH family protein | virB1 |
| FOB41_RS02455 (FOB41_02470) | 200703..201839 | - | 1137 | WP_234899609 | conjugal transfer protein TraB | - |
| FOB41_RS02460 (FOB41_02475) | 201862..202416 | - | 555 | WP_080823886 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 8382 | GenBank | NZ_CP050897 |
| Plasmid name | FDAARGOS_633|unnamed1 | Incompatibility group | - |
| Plasmid size | 208600 bp | Coordinate of oriT [Strand] | 205805..205841 [-] |
| Host baterium | Agrobacterium pusense strain FDAARGOS_633 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | dxnH |
| Symbiosis gene | - |
| Anti-CRISPR | - |