Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107944 |
| Name | oriT_YIC4027|unnamed |
| Organism | Sinorhizobium alkalisoli strain YIC4027 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP034911 (333658..333686 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_YIC4027|unnamed
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file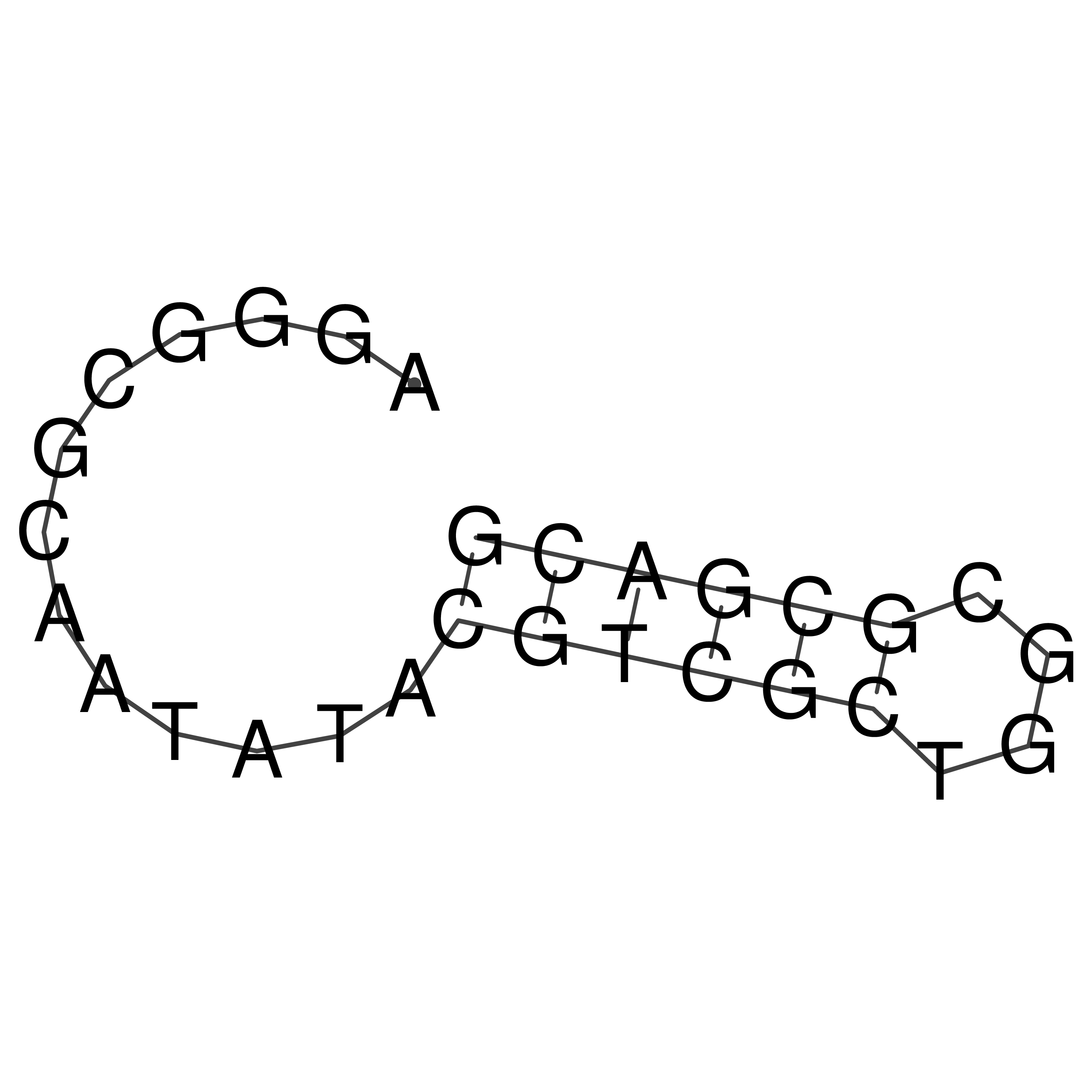
Relaxase
| ID | 5331 | GenBank | WP_069460873 |
| Name | traA_EKH55_RS28615_YIC4027|unnamed |
UniProt ID | A0A1E3V5C0 |
| Length | 1538 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1538 a.a. Molecular weight: 170233.49 Da Isoelectric Point: 9.5172
>WP_069460873.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium alkalisoli]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAAELMHEELALPEDIPAWLKAALDG
QSVAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSRGMVADWVYHDKDG
NAHIHLITTLRPLTEEGFGPKKVAVTGEDGEPLRVVTPDRSNGKIVYRLWAGDKETMKSWKIAWAETANR
HLALAGHEVRLDGRSYAEQGLDGIAQQHLGPEKAALARKGVEMYFAPADLARRQEMADRLLVEPELLLKQ
LANERSTFDERDIAKALHRYVDDPTGFANIRARLMASDDLVLLRPQQVDGQYGKVSEPAIFTTRDILRIE
YDMAQSAEHLAKQKGFGVSPRHVSAAVASIETANPEKAFKLDAEQVDAVRHVTGDNGIAAVVGLAGAGKS
TLLASARVAWESEHRRVIGAALAGKAAEGLEDNSGIRSRTLASWELAWAGGRELLHRGDVLVIDEAGMVS
SGQMARVLKTVEDAGAKAVLVGDAMQLQPIQAGAAFRAISERIGFAELAGVRRQRDEWARDASRLFARGN
VERGLDAYAQRGHITEAERRDEIIGRIVADWTEARRDLLQKITDGGTAPRLRGDELLVLAHTNEDVKRLN
QSLRKVMMEESALTDAREYQTARGLREFAAGDRIIFLENARFLEPRARRHGPQYVKNGMLGTVVSTGDKQ
GDTLLTVRLDNGRDVVLSEDSYRNIDHGYAATIHKSQGATVDRTFILATGMMDQHLTYVAMTRHRDRADL
YAAREDFEGKPGWGHRPRVDHAAGVTGELVATGFARFRPEDEDSENPFADVRKNDGTVHRLWGVSLPKAL
KEAVVSDGDTVTLRKDGVEKVKVTIPVIDENTGEKLFEEREVDRNVWTAKLIESAKARVERIERENHRPE
LFGELVERLGRSGAKTTTLEFESEEGYRTLRQDFARRRGIDTLAGVAAGMEEGVSRQLAWIAEKREKVAK
LWERASVALGFAIAHERRTAYTEEPRVSLASDIPATGKYLIPPTTMFVRSVNEDARRAQLASPRWKERDA
ILRPVLKKIHRDPEAALVRLNTLVSDTMIEPRKLVDDLGVKPERLGRLHGSNLMVDGRAERDKRAAATAA
IKDLLPLARAHATEFCRTAGQFIERERTRRKYMAQSIPALSKPAMARLVEIEAIRERGGEDAYKTAFAYA
GEDRALVQEVKAVNDALTARFGWSAFTAKADAVAERNITDRMPDVLSPERRDKLTRLFAVIRRFAEEQHL
AEKQDRSKIVAGASVAAAPETVTVLPMLAAVTEFKTPVEEEARIRAIAAAHYRHHRAAFAETAMRVWRDP
AGGIEKIEALLAKGFAGDRIAEAVNNDPSAYGALRGSGRLVDKLLAPGRERKEALLAVPEAASRIRSLGA
SYASALDAEIRGIVEERRRMAVAIPGLSKAAEDALYRLTAEMSKKGSKLDVAAGSLDPAIAREFAIVSRA
LDERFGPKAILRGEKDVINRVAPAQRRAFEAMREKMKVLQQTVRHQGSEQVLSERRLRTIDRARGLSR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAAELMHEELALPEDIPAWLKAALDG
QSVAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSRGMVADWVYHDKDG
NAHIHLITTLRPLTEEGFGPKKVAVTGEDGEPLRVVTPDRSNGKIVYRLWAGDKETMKSWKIAWAETANR
HLALAGHEVRLDGRSYAEQGLDGIAQQHLGPEKAALARKGVEMYFAPADLARRQEMADRLLVEPELLLKQ
LANERSTFDERDIAKALHRYVDDPTGFANIRARLMASDDLVLLRPQQVDGQYGKVSEPAIFTTRDILRIE
YDMAQSAEHLAKQKGFGVSPRHVSAAVASIETANPEKAFKLDAEQVDAVRHVTGDNGIAAVVGLAGAGKS
TLLASARVAWESEHRRVIGAALAGKAAEGLEDNSGIRSRTLASWELAWAGGRELLHRGDVLVIDEAGMVS
SGQMARVLKTVEDAGAKAVLVGDAMQLQPIQAGAAFRAISERIGFAELAGVRRQRDEWARDASRLFARGN
VERGLDAYAQRGHITEAERRDEIIGRIVADWTEARRDLLQKITDGGTAPRLRGDELLVLAHTNEDVKRLN
QSLRKVMMEESALTDAREYQTARGLREFAAGDRIIFLENARFLEPRARRHGPQYVKNGMLGTVVSTGDKQ
GDTLLTVRLDNGRDVVLSEDSYRNIDHGYAATIHKSQGATVDRTFILATGMMDQHLTYVAMTRHRDRADL
YAAREDFEGKPGWGHRPRVDHAAGVTGELVATGFARFRPEDEDSENPFADVRKNDGTVHRLWGVSLPKAL
KEAVVSDGDTVTLRKDGVEKVKVTIPVIDENTGEKLFEEREVDRNVWTAKLIESAKARVERIERENHRPE
LFGELVERLGRSGAKTTTLEFESEEGYRTLRQDFARRRGIDTLAGVAAGMEEGVSRQLAWIAEKREKVAK
LWERASVALGFAIAHERRTAYTEEPRVSLASDIPATGKYLIPPTTMFVRSVNEDARRAQLASPRWKERDA
ILRPVLKKIHRDPEAALVRLNTLVSDTMIEPRKLVDDLGVKPERLGRLHGSNLMVDGRAERDKRAAATAA
IKDLLPLARAHATEFCRTAGQFIERERTRRKYMAQSIPALSKPAMARLVEIEAIRERGGEDAYKTAFAYA
GEDRALVQEVKAVNDALTARFGWSAFTAKADAVAERNITDRMPDVLSPERRDKLTRLFAVIRRFAEEQHL
AEKQDRSKIVAGASVAAAPETVTVLPMLAAVTEFKTPVEEEARIRAIAAAHYRHHRAAFAETAMRVWRDP
AGGIEKIEALLAKGFAGDRIAEAVNNDPSAYGALRGSGRLVDKLLAPGRERKEALLAVPEAASRIRSLGA
SYASALDAEIRGIVEERRRMAVAIPGLSKAAEDALYRLTAEMSKKGSKLDVAAGSLDPAIAREFAIVSRA
LDERFGPKAILRGEKDVINRVAPAQRRAFEAMREKMKVLQQTVRHQGSEQVLSERRLRTIDRARGLSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5600 | GenBank | WP_151614005 |
| Name | traG_EKH55_RS28630_YIC4027|unnamed |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 70380.30 Da Isoelectric Point: 9.6752
>WP_151614005.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium alkalisoli]
MALKWKLHPSLLLIFVPVAVTAFAVYVTGWRWQGLTAGLSGKAEYWFLRTAPLPALLFGPLSGLLTVWAL
PMHRRRPIAIASLACFLTVAGFYGLREFGRLSPFVKSGAVTWERALSFLDMVAVLGTVTAFMAVVVAARI
STVVPAPVKRARHGTFGDADWLPMSAAARVFPPDGEIVVGERYRVDREIVHKLPFDPNDPATWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSGPLICLDPSTEVAPMVVEHRTSGLGREVMVLDPT
NPVMGFNVLDGIETSKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYEGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKA
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGQAPLRCGRAIYFRRKDMHAAAK
RNRFVKN
MALKWKLHPSLLLIFVPVAVTAFAVYVTGWRWQGLTAGLSGKAEYWFLRTAPLPALLFGPLSGLLTVWAL
PMHRRRPIAIASLACFLTVAGFYGLREFGRLSPFVKSGAVTWERALSFLDMVAVLGTVTAFMAVVVAARI
STVVPAPVKRARHGTFGDADWLPMSAAARVFPPDGEIVVGERYRVDREIVHKLPFDPNDPATWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSGPLICLDPSTEVAPMVVEHRTSGLGREVMVLDPT
NPVMGFNVLDGIETSKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYEGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKA
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGQAPLRCGRAIYFRRKDMHAAAK
RNRFVKN
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 358469..368021
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EKH55_RS28725 (EKH55_5914) | 353609..354046 | - | 438 | WP_069460891 | hypothetical protein | - |
| EKH55_RS28730 (EKH55_5915) | 354120..356255 | - | 2136 | WP_151614006 | ParB/RepB/Spo0J family partition protein | - |
| EKH55_RS28735 (EKH55_5916) | 356494..356826 | + | 333 | WP_069460894 | integration host factor subunit beta | - |
| EKH55_RS28740 (EKH55_5917) | 357458..357814 | + | 357 | WP_069460895 | hypothetical protein | - |
| EKH55_RS28745 (EKH55_5918) | 357811..358398 | + | 588 | WP_069460896 | AAA family ATPase | - |
| EKH55_RS28750 (EKH55_5919) | 358469..359491 | - | 1023 | WP_069460897 | P-type DNA transfer ATPase VirB11 | virB11 |
| EKH55_RS28755 (EKH55_5920) | 359500..360672 | - | 1173 | WP_069460898 | type IV secretion system protein VirB10 | virB10 |
| EKH55_RS28760 (EKH55_5921) | 360688..361539 | - | 852 | WP_069460899 | P-type conjugative transfer protein VirB9 | virB9 |
| EKH55_RS28765 (EKH55_5922) | 361536..362207 | - | 672 | WP_069460900 | virB8 family protein | virB8 |
| EKH55_RS28770 | 362209..362466 | - | 258 | WP_069460901 | hypothetical protein | - |
| EKH55_RS28775 (EKH55_5924) | 362501..363433 | - | 933 | WP_069460902 | type IV secretion system protein | virB6 |
| EKH55_RS28780 (EKH55_5925) | 363437..363670 | - | 234 | WP_069460903 | EexN family lipoprotein | - |
| EKH55_RS28785 (EKH55_5926) | 363667..364359 | - | 693 | WP_245314752 | P-type DNA transfer protein VirB5 | virB5 |
| EKH55_RS28790 (EKH55_5927) | 364365..366731 | - | 2367 | WP_069460905 | VirB4 family type IV secretion system protein | virb4 |
| EKH55_RS28795 (EKH55_5928) | 366724..367062 | - | 339 | WP_069460906 | type IV secretion system protein VirB3 | virB3 |
| EKH55_RS28800 (EKH55_5929) | 367066..367365 | - | 300 | WP_069460907 | TrbC/VirB2 family protein | virB2 |
| EKH55_RS28805 (EKH55_5930) | 367362..368021 | - | 660 | WP_151614007 | lytic transglycosylase domain-containing protein | virB1 |
| EKH55_RS28810 (EKH55_5931) | 368024..368563 | - | 540 | WP_069460908 | hypothetical protein | - |
| EKH55_RS28815 (EKH55_5932) | 368665..369018 | + | 354 | WP_069460909 | MarR family transcriptional regulator | - |
| EKH55_RS28820 (EKH55_5933) | 369184..370086 | + | 903 | WP_069460910 | hypothetical protein | - |
| EKH55_RS28825 (EKH55_5934) | 370150..371121 | + | 972 | WP_069460911 | quinolinate synthase NadA | - |
| EKH55_RS28830 (EKH55_5935) | 371118..372413 | + | 1296 | Protein_367 | L-aspartate oxidase | - |
Host bacterium
| ID | 8379 | GenBank | NZ_CP034911 |
| Plasmid name | YIC4027|unnamed | Incompatibility group | - |
| Plasmid size | 456424 bp | Coordinate of oriT [Strand] | 333658..333686 [-] |
| Host baterium | Sinorhizobium alkalisoli strain YIC4027 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodJ, nodI, nodU, nodC, nodA, nodE, nodD, nolX, nolW, nolT, nolU, nolV, nodZ, nifX, nifN, nifE, nifD, nifH, nifS, fixA, fixB, fixC, fixX, nifB, nifZ, fixU |
| Anti-CRISPR | - |