Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107877 |
| Name | oriT_pJBIWA001_1 |
| Organism | Raoultella planticola strain JBIWA001 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP074183 (93519..93568 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pJBIWA001_1
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file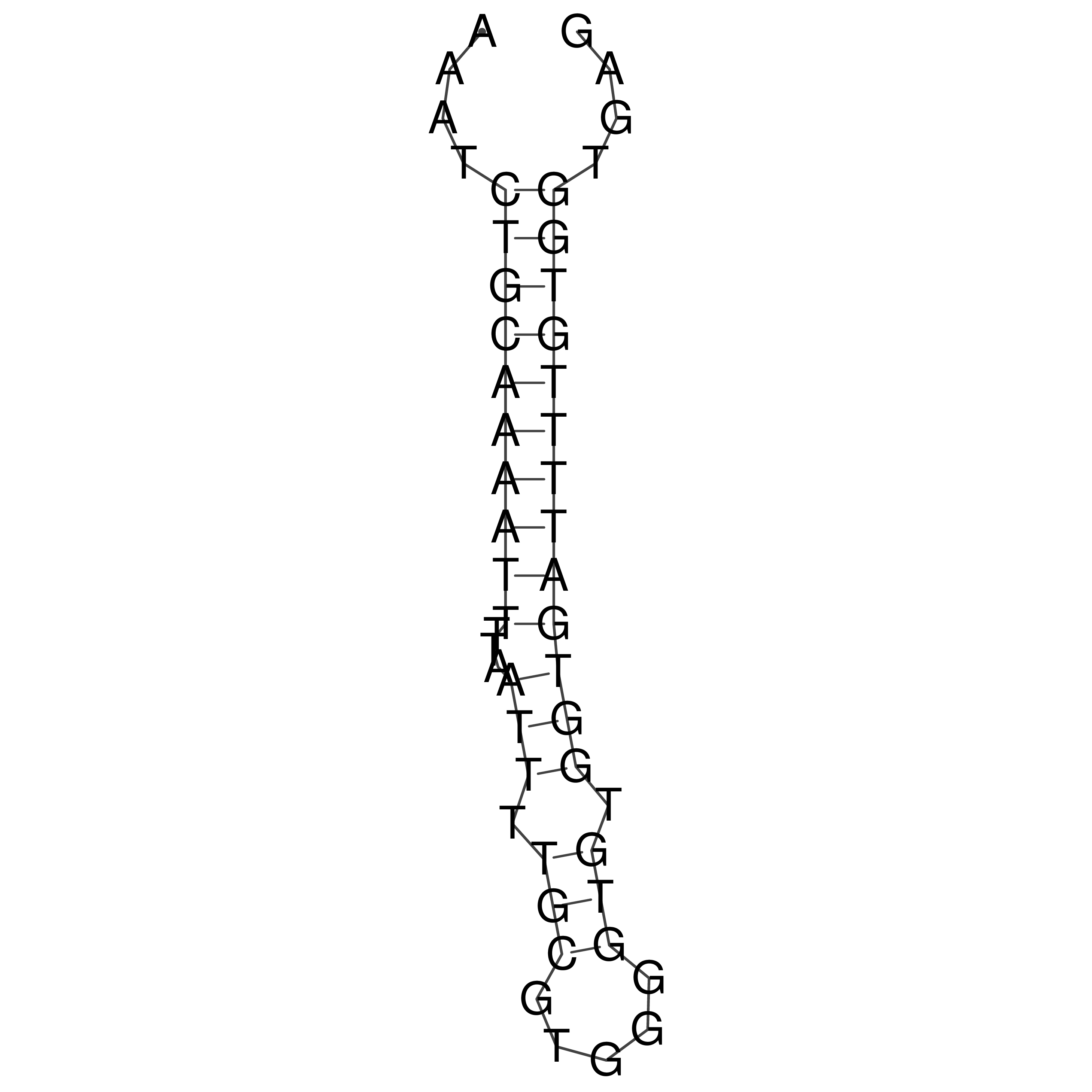
Relaxase
| ID | 5288 | GenBank | WP_223526237 |
| Name | traI_KGP19_RS27425_pJBIWA001_1 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190316.25 Da Isoelectric Point: 6.2703
>WP_223526237.1 conjugative transfer relaxase/helicase TraI [Raoultella planticola]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAANARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTQAQEKAIQ
KALSEGETLPAGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDIQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASEGLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAANARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTQAQEKAIQ
KALSEGETLPAGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDIQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASEGLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5542 | GenBank | WP_032072078 |
| Name | traD_KGP19_RS27420_pJBIWA001_1 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85822.78 Da Isoelectric Point: 4.9615
>WP_032072078.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 92961..121957
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KGP19_RS27210 (KGP19_27120) | 88063..88413 | + | 351 | WP_004153414 | hypothetical protein | - |
| KGP19_RS27215 (KGP19_27125) | 89043..89396 | + | 354 | WP_023179962 | hypothetical protein | - |
| KGP19_RS27220 (KGP19_27130) | 89453..89800 | + | 348 | WP_004152749 | hypothetical protein | - |
| KGP19_RS27225 (KGP19_27135) | 89895..90041 | + | 147 | WP_004152750 | hypothetical protein | - |
| KGP19_RS27230 (KGP19_27140) | 90092..90925 | + | 834 | WP_004152751 | N-6 DNA methylase | - |
| KGP19_RS27235 (KGP19_27145) | 91745..92566 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| KGP19_RS27240 (KGP19_27150) | 92599..92928 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| KGP19_RS27245 (KGP19_27155) | 92961..93446 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| KGP19_RS27250 (KGP19_27160) | 93868..94266 | + | 399 | WP_004152493 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| KGP19_RS27255 (KGP19_27165) | 94440..95126 | + | 687 | WP_004152494 | transcriptional regulator TraJ family protein | - |
| KGP19_RS27260 (KGP19_27170) | 95205..95591 | + | 387 | WP_004152495 | TraY domain-containing protein | - |
| KGP19_RS27265 (KGP19_27175) | 95645..96013 | + | 369 | WP_004152496 | type IV conjugative transfer system pilin TraA | - |
| KGP19_RS27270 (KGP19_27180) | 96027..96332 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| KGP19_RS27275 (KGP19_27185) | 96352..96918 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| KGP19_RS27280 (KGP19_27190) | 96905..97645 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| KGP19_RS27285 (KGP19_27195) | 97645..99069 | + | 1425 | WP_223526234 | F-type conjugal transfer pilus assembly protein TraB | traB |
| KGP19_RS27290 (KGP19_27200) | 99183..99767 | + | 585 | WP_004161368 | type IV conjugative transfer system lipoprotein TraV | traV |
| KGP19_RS27295 (KGP19_27205) | 99899..100309 | + | 411 | WP_032329942 | hypothetical protein | - |
| KGP19_RS27300 (KGP19_27210) | 100415..100633 | + | 219 | WP_004152501 | hypothetical protein | - |
| KGP19_RS27305 (KGP19_27215) | 100634..100945 | + | 312 | WP_004152502 | hypothetical protein | - |
| KGP19_RS27310 (KGP19_27220) | 101012..101416 | + | 405 | WP_004152503 | hypothetical protein | - |
| KGP19_RS27315 (KGP19_27225) | 101459..101848 | + | 390 | WP_223526235 | hypothetical protein | - |
| KGP19_RS27320 (KGP19_27230) | 101856..102254 | + | 399 | WP_011977783 | hypothetical protein | - |
| KGP19_RS27325 (KGP19_27235) | 102326..104965 | + | 2640 | WP_004152505 | type IV secretion system protein TraC | virb4 |
| KGP19_RS27330 (KGP19_27240) | 104965..105354 | + | 390 | WP_004152506 | type-F conjugative transfer system protein TrbI | - |
| KGP19_RS27335 (KGP19_27245) | 105354..105980 | + | 627 | WP_032420974 | type-F conjugative transfer system protein TraW | traW |
| KGP19_RS27340 (KGP19_27250) | 106022..106411 | + | 390 | WP_004152508 | hypothetical protein | - |
| KGP19_RS27345 (KGP19_27255) | 106408..107397 | + | 990 | WP_011977785 | conjugal transfer pilus assembly protein TraU | traU |
| KGP19_RS27350 (KGP19_27260) | 107410..108048 | + | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| KGP19_RS27355 (KGP19_27265) | 108107..110062 | + | 1956 | WP_030003482 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| KGP19_RS27360 (KGP19_27270) | 110094..110348 | + | 255 | WP_223526236 | conjugal transfer protein TrbE | - |
| KGP19_RS27365 (KGP19_27275) | 110326..110574 | + | 249 | WP_004152675 | hypothetical protein | - |
| KGP19_RS27370 (KGP19_27280) | 110587..110913 | + | 327 | WP_125064279 | hypothetical protein | - |
| KGP19_RS27375 (KGP19_27285) | 110934..111686 | + | 753 | WP_074015674 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| KGP19_RS27380 (KGP19_27290) | 111697..111936 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| KGP19_RS27385 (KGP19_27295) | 111908..112465 | + | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| KGP19_RS27390 (KGP19_27300) | 112510..112953 | + | 444 | WP_015065638 | F-type conjugal transfer protein TrbF | - |
| KGP19_RS27395 (KGP19_27305) | 112931..114310 | + | 1380 | WP_004165169 | conjugal transfer pilus assembly protein TraH | traH |
| KGP19_RS27400 (KGP19_27310) | 114310..117159 | + | 2850 | WP_004152624 | conjugal transfer mating-pair stabilization protein TraG | traG |
| KGP19_RS27405 (KGP19_27315) | 117172..117720 | + | 549 | WP_004152623 | conjugal transfer entry exclusion protein TraS | - |
| KGP19_RS27410 (KGP19_27320) | 117904..118635 | + | 732 | WP_004152622 | conjugal transfer complement resistance protein TraT | - |
| KGP19_RS27415 (KGP19_27325) | 118827..119516 | + | 690 | WP_022644928 | hypothetical protein | - |
| KGP19_RS27420 (KGP19_27330) | 119645..121957 | + | 2313 | WP_032072078 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 8312 | GenBank | NZ_CP074183 |
| Plasmid name | pJBIWA001_1 | Incompatibility group | IncFIB |
| Plasmid size | 136413 bp | Coordinate of oriT [Strand] | 93519..93568 [-] |
| Host baterium | Raoultella planticola strain JBIWA001 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |