Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107833 |
| Name | oriT1_pEF45-5 |
| Organism | Escherichia fergusonii strain EF20JDJ4045 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP086609 (6779..6859 [-], 81 nt) |
| oriT length | 81 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 81 nt
>oriT1_pEF45-5
GGGGTGCCGGGGCAAAGCCCTGACCAGACAGCAAAAGGAATATCGTCTCGGGTGACGGTATTACATTTGAACATTCTGTCC
GGGGTGCCGGGGCAAAGCCCTGACCAGACAGCAAAAGGAATATCGTCTCGGGTGACGGTATTACATTTGAACATTCTGTCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file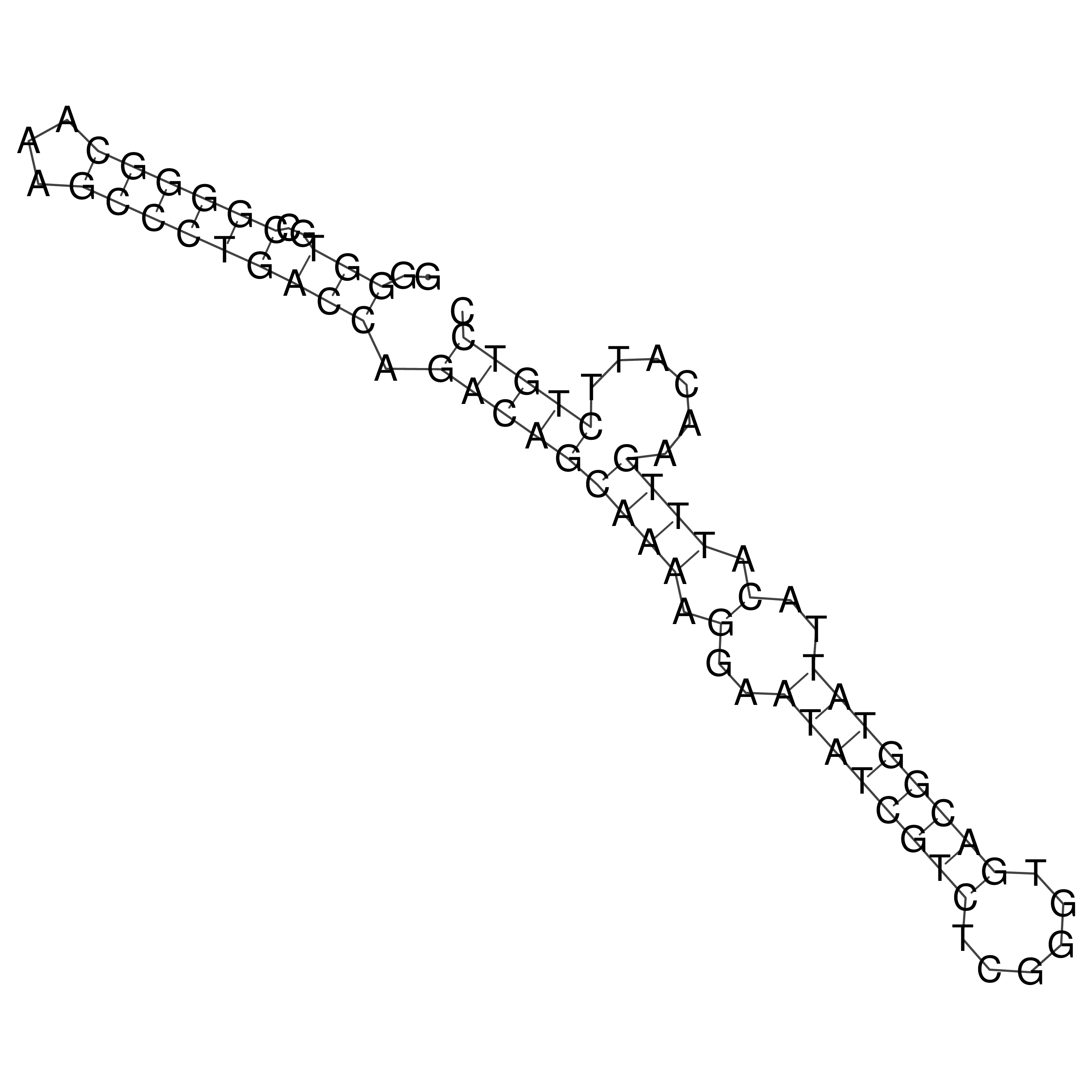
Relaxase
| ID | 5257 | GenBank | WP_125401188 |
| Name | nikB_LNN29_RS25110_pEF45-5 |
UniProt ID | _ |
| Length | 902 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 902 a.a. Molecular weight: 104384.85 Da Isoelectric Point: 7.6237
>WP_125401188.1 IncI1-type relaxase NikB [Escherichia fergusonii]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHPGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPAERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPVLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVRGRTGHEDYRITRPDVDHHREGSERHYRDYIAANSNDDASLPPPEQDKRREQPSPVLSR
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHPGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPAERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPVLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVRGRTGHEDYRITRPDVDHHREGSERHYRDYIAANSNDDASLPPPEQDKRREQPSPVLSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 2408 | GenBank | WP_001284001 |
| Name | WP_001284001_pEF45-5 |
UniProt ID | A0A5T9R3M9 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12599.50 Da Isoelectric Point: 10.8113
>WP_001284001.1 MULTISPECIES: IncI1-type relaxosome accessory protein NikA [Enterobacteriaceae]
MSDSGVRKKSEVRQKTVVRTLRFSPAEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDREYAEVLVAITELTNTLRKQLMEG
MSDSGVRKKSEVRQKTVVRTLRFSPAEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDREYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5T9R3M9 |
Host bacterium
| ID | 8268 | GenBank | NZ_CP086609 |
| Plasmid name | pEF45-5 | Incompatibility group | IncX1 |
| Plasmid size | 46074 bp | Coordinate of oriT [Strand] | 6779..6859 [-]; 30145..30232 [+] |
| Host baterium | Escherichia fergusonii strain EF20JDJ4045 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |