Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107827 |
| Name | oriT_FAM24678|unnamed1 |
| Organism | Morganella morganii strain FAM24678 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP066131 (6977..7012 [-], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 9..16, 18..25 (GCAAATTT..AAATTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_FAM24678|unnamed1
AAATTCATGCAAATTTTAAATTTGCGTAGTGTGTGG
AAATTCATGCAAATTTTAAATTTGCGTAGTGTGTGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file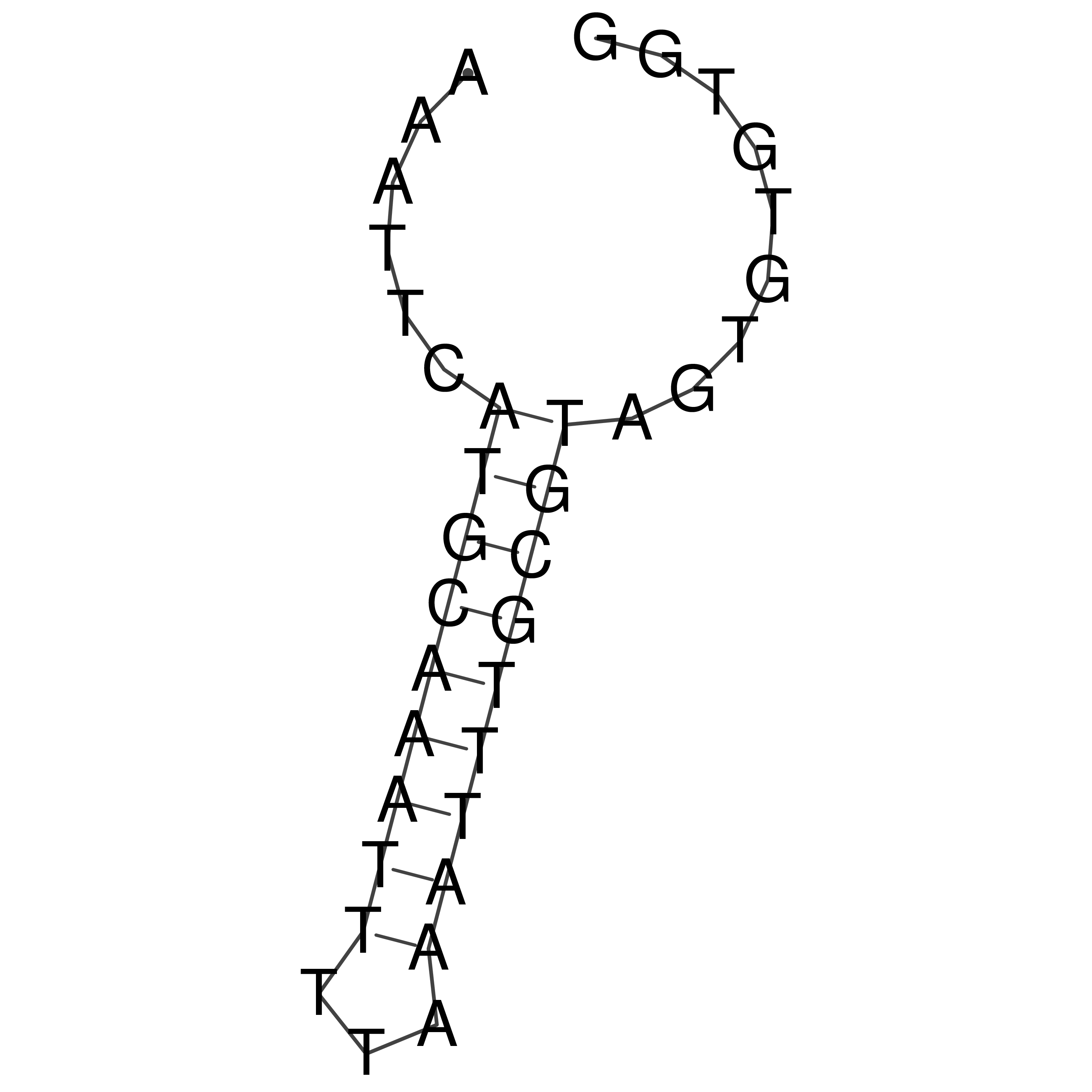
Relaxase
| ID | 5253 | GenBank | WP_096862173 |
| Name | mobF_JC792_RS20000_FAM24678|unnamed1 |
UniProt ID | _ |
| Length | 1693 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1693 a.a. Molecular weight: 184635.03 Da Isoelectric Point: 6.9602
>WP_096862173.1 MobF family relaxase [Morganella morganii]
MLSVAPIAQNAENYYTAQDNYYFLGNAESRWLGKGAEALGLTGEVSKEQLREILAGRLPEGSSLERTENG
KNTHRQGHDLTFSAPKSVSVLAVVLGDERMLAAHNAAVTAALKETETLASTRVMKDGISAPVMTGNLVIA
AFNHDTSRELDPQLHTHALVMNMTAHEGQWRTLSSDTRHNSGFSEAVYKTRISLGRIYLHELRQQVEKMG
FKTHDTGRNGLWEIEGIPVAPFSQRRQQILDAAGHDASLKSRDVAALDTRQAKRTPDKAELLTEWYDRLD
KNGFGSEERSRFNTAAQSREQDSLTPPPLPAALQKAADAALTEAVNALSERQVSMTYSAVLAKTLSGLDA
RPGIITVVRAAIDRAIEQRQLIPLDEQKGLFTSAAHLTDELTLQQLAAEVRTENRTPVFRVPVTPFTGTA
QKLADTLPNAAVIDSRGNRQMQRDRLAETAAVALLQGRTVLVIEADSKHTPPPVSGTLSTDKRRWTEKPA
LPVNGTVIVTGAETWSLKETVQLFAQAKETNTQILLLDSAGRKGTGNALRTLEDAGVVRFTAAGKPPVQT
EIHSIGDKQQRYAAVAARYQALRREGVPVTVQVTGTRERAALTDAVRAELTAQGVLQGSPVSVTLLTPVW
SDSKTRRDISQYREGMILEQWQDGKKSAERFTVSRVTPATRSVTLEDSRGQSQVIKVTKLDSSWSLFHPA
QKAVAAGETLTLLAKHGRMAAGDTLTVTAVTDSILTAEHRGRRVTLPLTAGLKADYAYVTTPGQSLPDNQ
VVLAAASVRDTRADFFNTVARSGEKIELFTPLDNTETDRRLAKSPHYQTVAARLGTDPQNPDNSVTQAAE
RLWSVPEKAVRQGIVLTQENQVMFSVTDVVAKTLPLHPSLTAESIRLQTAKMAAAGELIPVPGSGKNEYV
TAAAWESEKQIIRAILQGKGTQQPFFERVPEAMLTGLTPGQQAATRLILESTDRFIAVQGYAGVGKTTQF
KAVMRSLDLLPEDARPEVRGLAPTHRAVGEMAATGVKAQTLQSFLTDTAQQLRNGETPRFDNTLFLIDES
SMIGNRNMAEVVAVIEAGGGRAVKSGDTAQEQAIDSGTPFELAQTRSALDTAIMKEIVRQTPALKPAVEA
VIAGQMKQAAALLENVSPDIVPRLPDAHVPEKSVMQAGEEGVIRDIVRDYSSRTPEAREKTLIVVQTNAD
RAAVNAGVHEALTAGQDVRRIRIPVLVQEKTQTESLHSVAGLAQHHGKTVLAGDNYYTLIVTERDKADGV
VMLRDADKREFPLSAFENSSRDIQVFRTETREMAEGEKVSFTRTDRNRGRVVNSTRTVSQIHDDGTLTLT
DGRDSIRINPAEPADRHIDYGYAGTAHKAQGASEEYVIVLGGVTGGRQPLASRRNAYVALSRMKTHVQVY
SDDLDKWLDKTGSNAVRPTVHDLLLADSDRAAATGNRLYAGAKPLSDTAVGRTLSRELGLTAESTARFVY
GSQKYPEPGVAWPVYDRHGRPAGTRIMPVLLNEQGHLNGAGQEHRLVGREEAAWMVLQRGRNGETRIAES
MTEALAQLKQTPESGVVVRLQPEEPVNAAVMQKLTGGLPADLSVPDTLRAGVPAADDPMTLKTAEEQRRE
AELKAAALPSKTDTVRDAPQDARLEAEITREQQRLQSQENAHEQKAAISLDDMIRQEQLHRMRQTEQLQQ
AEKDIVREKEIGE
MLSVAPIAQNAENYYTAQDNYYFLGNAESRWLGKGAEALGLTGEVSKEQLREILAGRLPEGSSLERTENG
KNTHRQGHDLTFSAPKSVSVLAVVLGDERMLAAHNAAVTAALKETETLASTRVMKDGISAPVMTGNLVIA
AFNHDTSRELDPQLHTHALVMNMTAHEGQWRTLSSDTRHNSGFSEAVYKTRISLGRIYLHELRQQVEKMG
FKTHDTGRNGLWEIEGIPVAPFSQRRQQILDAAGHDASLKSRDVAALDTRQAKRTPDKAELLTEWYDRLD
KNGFGSEERSRFNTAAQSREQDSLTPPPLPAALQKAADAALTEAVNALSERQVSMTYSAVLAKTLSGLDA
RPGIITVVRAAIDRAIEQRQLIPLDEQKGLFTSAAHLTDELTLQQLAAEVRTENRTPVFRVPVTPFTGTA
QKLADTLPNAAVIDSRGNRQMQRDRLAETAAVALLQGRTVLVIEADSKHTPPPVSGTLSTDKRRWTEKPA
LPVNGTVIVTGAETWSLKETVQLFAQAKETNTQILLLDSAGRKGTGNALRTLEDAGVVRFTAAGKPPVQT
EIHSIGDKQQRYAAVAARYQALRREGVPVTVQVTGTRERAALTDAVRAELTAQGVLQGSPVSVTLLTPVW
SDSKTRRDISQYREGMILEQWQDGKKSAERFTVSRVTPATRSVTLEDSRGQSQVIKVTKLDSSWSLFHPA
QKAVAAGETLTLLAKHGRMAAGDTLTVTAVTDSILTAEHRGRRVTLPLTAGLKADYAYVTTPGQSLPDNQ
VVLAAASVRDTRADFFNTVARSGEKIELFTPLDNTETDRRLAKSPHYQTVAARLGTDPQNPDNSVTQAAE
RLWSVPEKAVRQGIVLTQENQVMFSVTDVVAKTLPLHPSLTAESIRLQTAKMAAAGELIPVPGSGKNEYV
TAAAWESEKQIIRAILQGKGTQQPFFERVPEAMLTGLTPGQQAATRLILESTDRFIAVQGYAGVGKTTQF
KAVMRSLDLLPEDARPEVRGLAPTHRAVGEMAATGVKAQTLQSFLTDTAQQLRNGETPRFDNTLFLIDES
SMIGNRNMAEVVAVIEAGGGRAVKSGDTAQEQAIDSGTPFELAQTRSALDTAIMKEIVRQTPALKPAVEA
VIAGQMKQAAALLENVSPDIVPRLPDAHVPEKSVMQAGEEGVIRDIVRDYSSRTPEAREKTLIVVQTNAD
RAAVNAGVHEALTAGQDVRRIRIPVLVQEKTQTESLHSVAGLAQHHGKTVLAGDNYYTLIVTERDKADGV
VMLRDADKREFPLSAFENSSRDIQVFRTETREMAEGEKVSFTRTDRNRGRVVNSTRTVSQIHDDGTLTLT
DGRDSIRINPAEPADRHIDYGYAGTAHKAQGASEEYVIVLGGVTGGRQPLASRRNAYVALSRMKTHVQVY
SDDLDKWLDKTGSNAVRPTVHDLLLADSDRAAATGNRLYAGAKPLSDTAVGRTLSRELGLTAESTARFVY
GSQKYPEPGVAWPVYDRHGRPAGTRIMPVLLNEQGHLNGAGQEHRLVGREEAAWMVLQRGRNGETRIAES
MTEALAQLKQTPESGVVVRLQPEEPVNAAVMQKLTGGLPADLSVPDTLRAGVPAADDPMTLKTAEEQRRE
AELKAAALPSKTDTVRDAPQDARLEAEITREQQRLQSQENAHEQKAAISLDDMIRQEQLHRMRQTEQLQQ
AEKDIVREKEIGE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5499 | GenBank | WP_096862174 |
| Name | traD_JC792_RS19995_FAM24678|unnamed1 |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 83586.48 Da Isoelectric Point: 5.2300
>WP_096862174.1 type IV conjugative transfer system coupling protein TraD [Morganella morganii]
MSSTKHITQGGQVFGYMLGMFMQVNKRISFYLILFYLILTPVIFYLKVPLQEIKNGGLYWWLYFMSPGEK
ALYRVPPVYDFNFDGTVYHFTSANILKDPYMTYAGDIFLQELWLSLFWSSVLVLLLAFVVYSLLHRLGEK
QARDETTGGRELTEDVSAVARQMQRRREASPIRIDKLPLILNSEVKNMLMHGTPGSGKSNTINKLLLQLR
ERGDMVIIFDKGCSLVKKHFNENCDKLLNPLDARCENWDLWRECLTTPDFDSMSNTLIPQGTKEDPFWTG
SARTIFTAVAAKLRNDPERSYNKLLRTLLAIDLKALREYVAGTEASNLMEEKVEKTAISIRGVLTNYVKA
LRYLQGIERNGKAPFAIREWMKQANEPKKPHGWLWITSNARQHESLKPLISMWLAQAANCVLEMNENPER
RIWFIYDELASLHKLPELPQVLSEARKFGGCFVLGFQNKPQLDFTYGEDYADAMMDLLNTRFFFRSPDQN
VAMWVRDQLGQKRAKLFNEQYSYGADTVRDGVSFTKQEDDKTLVSYSDIQKLPDLECYVTLPGDYPVVRM
KMTYEKIRPVAAEFIERELNDSLEQDITARLDAGEQDDARLQRLLSSSTPAPETPSGLPPQTAAVLVVPP
VLTMAAAHPAPVVPSVTTPVTAVSSENTTQTAQTDTAVTGGREQDVPDMVIDTATGEIVYPGADEPDAET
RAVMYENYSALHQEEKNIAVHRRAEPDDHDEPEVW
MSSTKHITQGGQVFGYMLGMFMQVNKRISFYLILFYLILTPVIFYLKVPLQEIKNGGLYWWLYFMSPGEK
ALYRVPPVYDFNFDGTVYHFTSANILKDPYMTYAGDIFLQELWLSLFWSSVLVLLLAFVVYSLLHRLGEK
QARDETTGGRELTEDVSAVARQMQRRREASPIRIDKLPLILNSEVKNMLMHGTPGSGKSNTINKLLLQLR
ERGDMVIIFDKGCSLVKKHFNENCDKLLNPLDARCENWDLWRECLTTPDFDSMSNTLIPQGTKEDPFWTG
SARTIFTAVAAKLRNDPERSYNKLLRTLLAIDLKALREYVAGTEASNLMEEKVEKTAISIRGVLTNYVKA
LRYLQGIERNGKAPFAIREWMKQANEPKKPHGWLWITSNARQHESLKPLISMWLAQAANCVLEMNENPER
RIWFIYDELASLHKLPELPQVLSEARKFGGCFVLGFQNKPQLDFTYGEDYADAMMDLLNTRFFFRSPDQN
VAMWVRDQLGQKRAKLFNEQYSYGADTVRDGVSFTKQEDDKTLVSYSDIQKLPDLECYVTLPGDYPVVRM
KMTYEKIRPVAAEFIERELNDSLEQDITARLDAGEQDDARLQRLLSSSTPAPETPSGLPPQTAAVLVVPP
VLTMAAAHPAPVVPSVTTPVTAVSSENTTQTAQTDTAVTGGREQDVPDMVIDTATGEIVYPGADEPDAET
RAVMYENYSALHQEEKNIAVHRRAEPDDHDEPEVW
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 9524..32262
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JC792_RS19840 (JC792_19835) | 5221..6165 | + | 945 | WP_218453978 | zincin-like metallopeptidase domain-containing protein | - |
| JC792_RS19845 (JC792_19840) | 6447..6761 | - | 315 | WP_348773190 | lytic transglycosylase domain-containing protein | - |
| JC792_RS19850 (JC792_19845) | 7320..7703 | + | 384 | WP_218453979 | relaxosome protein TraM | - |
| JC792_RS19855 (JC792_19850) | 7906..8625 | + | 720 | WP_218453980 | hypothetical protein | - |
| JC792_RS19860 (JC792_19855) | 8677..8871 | + | 195 | WP_218453981 | TraY domain-containing protein | - |
| JC792_RS19865 (JC792_19860) | 9118..9495 | + | 378 | WP_172694096 | type IV conjugative transfer system pilin TraA | - |
| JC792_RS19870 (JC792_19865) | 9524..9829 | + | 306 | WP_172694066 | type IV conjugative transfer system protein TraL | traL |
| JC792_RS19875 (JC792_19870) | 9840..10412 | + | 573 | WP_218453982 | type IV conjugative transfer system protein TraE | traE |
| JC792_RS19880 (JC792_19875) | 10393..11166 | + | 774 | WP_247717512 | type-F conjugative transfer system secretin TraK | traK |
| JC792_RS19885 (JC792_19880) | 11168..12550 | + | 1383 | WP_218453983 | TrbI/VirB10 family protein | traB |
| JC792_RS19890 (JC792_19885) | 12566..12790 | + | 225 | WP_172694063 | hypothetical protein | - |
| JC792_RS19895 (JC792_19890) | 12792..12956 | + | 165 | WP_172694062 | hypothetical protein | - |
| JC792_RS19900 (JC792_19895) | 13113..13292 | + | 180 | WP_218453984 | hypothetical protein | - |
| JC792_RS19905 (JC792_19900) | 13303..13629 | + | 327 | WP_218453985 | hypothetical protein | - |
| JC792_RS19910 (JC792_19905) | 13754..14014 | + | 261 | WP_218453986 | hypothetical protein | - |
| JC792_RS19915 (JC792_19910) | 14019..14540 | + | 522 | WP_172694058 | type IV conjugative transfer system lipoprotein TraV | traV |
| JC792_RS19920 (JC792_19915) | 14540..17248 | + | 2709 | WP_247717513 | type IV secretion system protein TraC | virb4 |
| JC792_RS20575 | 17224..17625 | + | 402 | Protein_23 | type-F conjugative transfer system protein TrbI | - |
| JC792_RS20580 | 17653..18279 | + | 627 | WP_172694056 | type-F conjugative transfer system protein TraW | traW |
| JC792_RS19930 (JC792_19925) | 18291..19259 | + | 969 | WP_247715250 | conjugal transfer pilus assembly protein TraU | traU |
| JC792_RS19935 (JC792_19930) | 19271..19882 | + | 612 | WP_247717514 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| JC792_RS19940 (JC792_19935) | 19882..21699 | + | 1818 | WP_218453988 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| JC792_RS19945 (JC792_19940) | 21696..22475 | + | 780 | WP_218453989 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| JC792_RS19950 (JC792_19945) | 22491..22775 | + | 285 | WP_193830285 | hypothetical protein | - |
| JC792_RS19955 (JC792_19950) | 22772..23296 | + | 525 | WP_218453990 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| JC792_RS19960 (JC792_19955) | 23390..23746 | + | 357 | WP_130184729 | hypothetical protein | - |
| JC792_RS19965 (JC792_19960) | 23739..25136 | + | 1398 | WP_218453991 | conjugal transfer pilus assembly protein TraH | traH |
| JC792_RS19970 (JC792_19965) | 25126..27993 | + | 2868 | WP_218453992 | conjugal transfer mating-pair stabilization protein TraG | traG |
| JC792_RS19975 (JC792_19970) | 28050..28373 | + | 324 | WP_218453993 | YbjQ family protein | - |
| JC792_RS19980 (JC792_19975) | 28415..29152 | + | 738 | WP_218453994 | hypothetical protein | - |
| JC792_RS19985 (JC792_19980) | 29246..29452 | + | 207 | WP_096862176 | TraR/DksA C4-type zinc finger protein | - |
| JC792_RS19990 (JC792_19985) | 29516..29893 | + | 378 | WP_096862175 | hypothetical protein | - |
| JC792_RS19995 (JC792_19990) | 30055..32262 | + | 2208 | WP_096862174 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 8262 | GenBank | NZ_CP066131 |
| Plasmid name | FAM24678|unnamed1 | Incompatibility group | - |
| Plasmid size | 72755 bp | Coordinate of oriT [Strand] | 6977..7012 [-] |
| Host baterium | Morganella morganii strain FAM24678 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |