Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107804 |
| Name | oriT_pMM1L5 |
| Organism | Morganella morganii strain MM1L5 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MK851048 (60636..60670 [+], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 9..16, 18..25 (GCAAATTT..AAATTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
>oriT_pMM1L5
AAATTCATGCAAATTTTAAATTTGCGTAGTGTGTG
AAATTCATGCAAATTTTAAATTTGCGTAGTGTGTG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file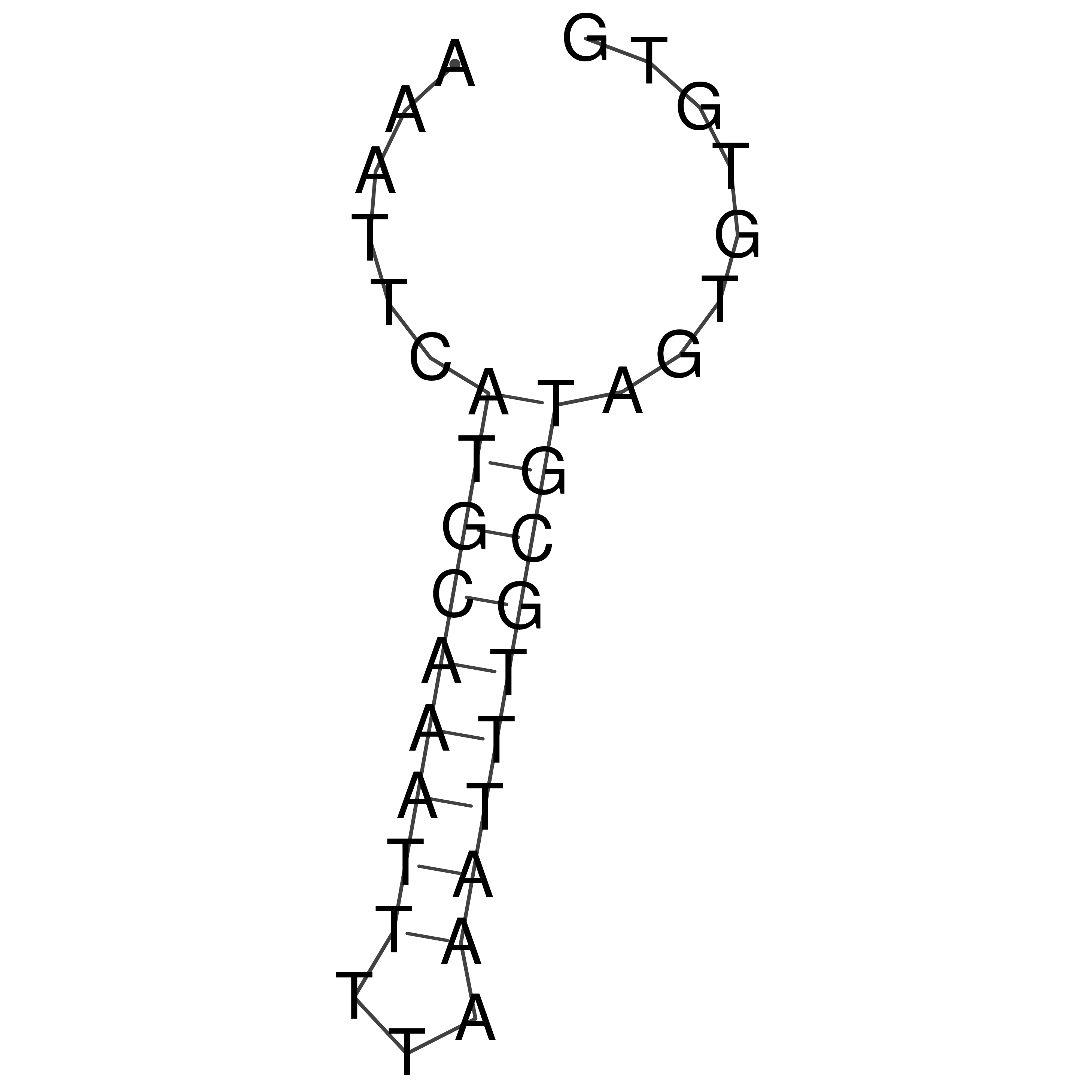
Relaxase
| ID | 5241 | GenBank | WP_172694085 |
| Name | traI_HTX04_RS00190_pMM1L5 |
UniProt ID | _ |
| Length | 1699 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1699 a.a. Molecular weight: 185311.71 Da Isoelectric Point: 7.0913
>WP_172694085.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Gammaproteobacteria]
MLSVAPIAKNAENYYTAQDNYYFLGNAESRWLGKGAEALGLTGEVSKEQLREILAGRLPEGSSLERTENG
KNTHRQGHDLTFSAPKSVSVLAVVLGDERMIAAHNDSVASALKEAETLASTRVMKDGQSVPVMTGNLVIA
AFNHDTSRELDPQVHTHGLVMNMTEYEGNWRTLSSDTRHNSGFSEAVYKLRISLGRIYLHRMRQKVEAMG
YKTYDSGPNGLWEIEGVPVAPFSQRHQQILDAAGHDASLKSRDVAALDTRRAKRTPDKEELLTEWFDRLD
KNGFGSEARSRFDAEAKSREQDSLTPPPLPAALQKVADAALTEAVNALSERQVSMTYSAVLAKTLSGLDA
RPGIITVVRNAIDRAIEQRQLIPLDEQKGLFTSAAHLTDELTLQQLAAEVRTENRTPVFRVPETVFTGTA
QKLADTLPNAAVIDSRGNRQMQRDRLAETAAVALQQGRTVLVIEADSKHTPEPVSGTLTTDKRRWTEKPA
LPVNGTVIVTGAETWSLKETVQLFAQAKETNTQILLLDSAGRKGTGNALRTLEDAGVVRFAAAGKPPVQT
EIHSIGDKQQRYAAVAARYQALRREGVPVTVQVTGTRERAALTDAVRAELTAQGVLHGSPVSVTLLTPVW
SDSKTRRDISQYREGMILEQWQDGKKSAERFTVSRVTPATRSVTLEDSRGQSQVIKVTKLDSSWSLFHPA
QKNIAAGETLTLLAKHGRLAAGDTLTVTAVTDSTLTAEHRGRRVTLPLTAGLKADYAYVTTPGQSLPDNQ
VVLAAASVRDTRADFFNTVARSGKKVELFTPLDNTETDRRLARSPHYQTVAARLGTDPQNPDTSVRQAAE
RLWSVPEKAVRQGIVLTQENQVMFSVTDVVAKTLPLHPSLTAESIRLQTAKMAAAGELIPVPGSGKNEYV
TAAAWESEKQIIRAILQGKGTQQPFFERVPESVLAGLTPGQQAATWLILESTDRFIAVQGYAGVGKTTQF
KAVMRSLDLLPEDARPEVRGLAPTHRAVGEMAATGVKAQTLQSFLTDTAQLLRNGETPRFDNTLFLIDES
SMIGNRNMAEVVAVIEAGGGRAVKSGDTAQEQAIDSGTPFELAQTRSALDTAIMKEIVRQTPALKPAVEA
VIAGQMKQAAALLENVSPDIVPRLPDAHIPEKSVMQAGEEGVIRDIVRDYSSRTPEAREKTLIVVQTNAD
RTAVNAGVHEALTAGADVRRVRIPVLVQEKTQTESLHSVAGLAQHHGKTVLAGDTYYTLIVTERDKADGV
VMLRDADKRDIPLSAFENSSRDIQVFRTETREMAEGEKVTFTRTDRNRGRVVNSTRTVSQIHDDGTLTLT
DGQGSIRINPAEPADRHIDYGYAGTAHKAQGASEEYVIVLGGVTGGRQPLASRRNAYVALSRMKTHVQVY
SDNLEKWLDKTGSNAARPTVHDLLLADSDRAAATGNRLYGAAKPLGDTAVGRALSRELGLTAESTARFVY
GSQKYPEPGVAWPVYDRHGRPAGTRITPVLLNEQGHLNGAGQEQRLVGREEAAWMVLQRGSNGETRIADS
MTEALAQLKQTPDSGVVVRLQPEEPVNAAVMRKLTGGLPADLSVPDMLRAGITAADDPITLKTAEEQRRE
AELKAAALPEKADTVRDASQDTRLDAEITREQQRLHSQENAHDRKAAVSLDEILRQEQLHRMRQTEKLQQ
AGKDIVREREPVREKEIGE
MLSVAPIAKNAENYYTAQDNYYFLGNAESRWLGKGAEALGLTGEVSKEQLREILAGRLPEGSSLERTENG
KNTHRQGHDLTFSAPKSVSVLAVVLGDERMIAAHNDSVASALKEAETLASTRVMKDGQSVPVMTGNLVIA
AFNHDTSRELDPQVHTHGLVMNMTEYEGNWRTLSSDTRHNSGFSEAVYKLRISLGRIYLHRMRQKVEAMG
YKTYDSGPNGLWEIEGVPVAPFSQRHQQILDAAGHDASLKSRDVAALDTRRAKRTPDKEELLTEWFDRLD
KNGFGSEARSRFDAEAKSREQDSLTPPPLPAALQKVADAALTEAVNALSERQVSMTYSAVLAKTLSGLDA
RPGIITVVRNAIDRAIEQRQLIPLDEQKGLFTSAAHLTDELTLQQLAAEVRTENRTPVFRVPETVFTGTA
QKLADTLPNAAVIDSRGNRQMQRDRLAETAAVALQQGRTVLVIEADSKHTPEPVSGTLTTDKRRWTEKPA
LPVNGTVIVTGAETWSLKETVQLFAQAKETNTQILLLDSAGRKGTGNALRTLEDAGVVRFAAAGKPPVQT
EIHSIGDKQQRYAAVAARYQALRREGVPVTVQVTGTRERAALTDAVRAELTAQGVLHGSPVSVTLLTPVW
SDSKTRRDISQYREGMILEQWQDGKKSAERFTVSRVTPATRSVTLEDSRGQSQVIKVTKLDSSWSLFHPA
QKNIAAGETLTLLAKHGRLAAGDTLTVTAVTDSTLTAEHRGRRVTLPLTAGLKADYAYVTTPGQSLPDNQ
VVLAAASVRDTRADFFNTVARSGKKVELFTPLDNTETDRRLARSPHYQTVAARLGTDPQNPDTSVRQAAE
RLWSVPEKAVRQGIVLTQENQVMFSVTDVVAKTLPLHPSLTAESIRLQTAKMAAAGELIPVPGSGKNEYV
TAAAWESEKQIIRAILQGKGTQQPFFERVPESVLAGLTPGQQAATWLILESTDRFIAVQGYAGVGKTTQF
KAVMRSLDLLPEDARPEVRGLAPTHRAVGEMAATGVKAQTLQSFLTDTAQLLRNGETPRFDNTLFLIDES
SMIGNRNMAEVVAVIEAGGGRAVKSGDTAQEQAIDSGTPFELAQTRSALDTAIMKEIVRQTPALKPAVEA
VIAGQMKQAAALLENVSPDIVPRLPDAHIPEKSVMQAGEEGVIRDIVRDYSSRTPEAREKTLIVVQTNAD
RTAVNAGVHEALTAGADVRRVRIPVLVQEKTQTESLHSVAGLAQHHGKTVLAGDTYYTLIVTERDKADGV
VMLRDADKRDIPLSAFENSSRDIQVFRTETREMAEGEKVTFTRTDRNRGRVVNSTRTVSQIHDDGTLTLT
DGQGSIRINPAEPADRHIDYGYAGTAHKAQGASEEYVIVLGGVTGGRQPLASRRNAYVALSRMKTHVQVY
SDNLEKWLDKTGSNAARPTVHDLLLADSDRAAATGNRLYGAAKPLGDTAVGRALSRELGLTAESTARFVY
GSQKYPEPGVAWPVYDRHGRPAGTRITPVLLNEQGHLNGAGQEQRLVGREEAAWMVLQRGSNGETRIADS
MTEALAQLKQTPDSGVVVRLQPEEPVNAAVMRKLTGGLPADLSVPDMLRAGITAADDPITLKTAEEQRRE
AELKAAALPEKADTVRDASQDTRLDAEITREQQRLHSQENAHDRKAAVSLDEILRQEQLHRMRQTEKLQQ
AGKDIVREREPVREKEIGE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5486 | GenBank | WP_172694086 |
| Name | traD_HTX04_RS00195_pMM1L5 |
UniProt ID | _ |
| Length | 742 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 742 a.a. Molecular weight: 84244.09 Da Isoelectric Point: 5.3912
>WP_172694086.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MSSTKHITQGGQVFGYMLGMFMQVNKRISFYLILFYLILTPVIFYLKVPSQEIKNGGLYWWLYFMSPGEK
ALYRVPPVYDFNFDGTVYHFTSANILKDPYMIYAGDIFLQELWLSLFWSSVLVLLLAFVVYTLLHRLGEK
QARDETTGGRELTEDVSAVARQMQRRREASPIRIDKLPLILNSEVKNMLMHGTPGSGKSNTINKLLLQLR
ERGDMVIIFDKGCSLVKKHFNENRDKLLNPLDARCENWDLWRECLTTPDFDSMSNTLIPQGTKEDPFWTG
SARTIFTAVAAKLRNDPERSYNKLLRTLLAIDLKALREYVAGTEASNLMEEKVEKTAISIRGVLTNYVKA
LRYLQGIERNGKAPFAIREWMRQANEPKKPHGWLWITSNARQHESLKPLISMWLAQAANCVLEMNENPER
RIWFIYDELASLHKLPELPQVLSEARKFGGCFVLGFQNKPQLDFTYGEDYADAMMDLLNTRFFFRSPDQN
VAMWVRDQLGQKRAKLFNEQYSYGADTVRDGVSFSKQEDDKTLVSYSDIQKLPDLECYVTLPGDYPVVRM
KMTYEKIRPVAAEFIERALNDSLEQDITAQLDAGEQDDARLQRLISGSSTPAPETPSRLPVQTAAVLIVP
PVLTPAAGHPAPVVSAAVPSVTTPATSVTAENTAQTTQTDTAVTGGREQDVPDMVIDTATGEIVYPGADE
PDAETRAAMYENYSALHQEEKNIAVHRRAEPDDHDEHEPEVW
MSSTKHITQGGQVFGYMLGMFMQVNKRISFYLILFYLILTPVIFYLKVPSQEIKNGGLYWWLYFMSPGEK
ALYRVPPVYDFNFDGTVYHFTSANILKDPYMIYAGDIFLQELWLSLFWSSVLVLLLAFVVYTLLHRLGEK
QARDETTGGRELTEDVSAVARQMQRRREASPIRIDKLPLILNSEVKNMLMHGTPGSGKSNTINKLLLQLR
ERGDMVIIFDKGCSLVKKHFNENRDKLLNPLDARCENWDLWRECLTTPDFDSMSNTLIPQGTKEDPFWTG
SARTIFTAVAAKLRNDPERSYNKLLRTLLAIDLKALREYVAGTEASNLMEEKVEKTAISIRGVLTNYVKA
LRYLQGIERNGKAPFAIREWMRQANEPKKPHGWLWITSNARQHESLKPLISMWLAQAANCVLEMNENPER
RIWFIYDELASLHKLPELPQVLSEARKFGGCFVLGFQNKPQLDFTYGEDYADAMMDLLNTRFFFRSPDQN
VAMWVRDQLGQKRAKLFNEQYSYGADTVRDGVSFSKQEDDKTLVSYSDIQKLPDLECYVTLPGDYPVVRM
KMTYEKIRPVAAEFIERALNDSLEQDITAQLDAGEQDDARLQRLISGSSTPAPETPSRLPVQTAAVLIVP
PVLTPAAGHPAPVVSAAVPSVTTPATSVTAENTAQTTQTDTAVTGGREQDVPDMVIDTATGEIVYPGADE
PDAETRAAMYENYSALHQEEKNIAVHRRAEPDDHDEHEPEVW
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 36911..61247
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTX04_RS00195 | 36911..39139 | - | 2229 | WP_172694086 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HTX04_RS00200 | 39297..39503 | - | 207 | WP_172694087 | TraR/DksA C4-type zinc finger protein | - |
| HTX04_RS00205 | 39669..39923 | + | 255 | WP_172694088 | hypothetical protein | - |
| HTX04_RS00210 | 40005..42857 | - | 2853 | WP_172694089 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTX04_RS00215 | 42847..44244 | - | 1398 | WP_172694090 | conjugal transfer pilus assembly protein TraH | traH |
| HTX04_RS00220 | 44338..44862 | - | 525 | WP_053091309 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTX04_RS00225 | 44859..45143 | - | 285 | WP_049246541 | hypothetical protein | - |
| HTX04_RS00230 | 45159..45950 | - | 792 | WP_049246540 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTX04_RS00235 | 45947..47764 | - | 1818 | WP_172694091 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTX04_RS00240 | 47764..48375 | - | 612 | WP_247715241 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTX04_RS00245 | 48387..49355 | - | 969 | WP_247715250 | conjugal transfer pilus assembly protein TraU | traU |
| HTX04_RS00250 | 49367..49993 | - | 627 | WP_172694056 | type-F conjugative transfer system protein TraW | traW |
| HTX04_RS00255 | 49993..50421 | - | 429 | WP_172694057 | type-F conjugative transfer system protein TrbI | - |
| HTX04_RS00260 | 50397..53105 | - | 2709 | WP_247715242 | type IV secretion system protein TraC | virb4 |
| HTX04_RS00265 | 53105..53626 | - | 522 | WP_172694058 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTX04_RS00270 | 53631..53891 | - | 261 | WP_172694059 | hypothetical protein | - |
| HTX04_RS00275 | 54016..54330 | - | 315 | WP_172694060 | hypothetical protein | - |
| HTX04_RS00280 | 54354..54602 | - | 249 | WP_172694061 | hypothetical protein | - |
| HTX04_RS00285 | 54692..54856 | - | 165 | WP_172694062 | hypothetical protein | - |
| HTX04_RS00290 | 54858..55082 | - | 225 | WP_172694063 | hypothetical protein | - |
| HTX04_RS00295 | 55098..56480 | - | 1383 | WP_172694064 | TrbI/VirB10 family protein | traB |
| HTX04_RS00300 | 56482..57255 | - | 774 | WP_247715243 | type-F conjugative transfer system secretin TraK | traK |
| HTX04_RS00305 | 57236..57808 | - | 573 | WP_172694065 | type IV conjugative transfer system protein TraE | traE |
| HTX04_RS00310 | 57819..58124 | - | 306 | WP_172694066 | type IV conjugative transfer system protein TraL | traL |
| HTX04_RS00315 | 58153..58530 | - | 378 | WP_172694096 | type IV conjugative transfer system pilin TraA | - |
| HTX04_RS00320 | 58777..58965 | - | 189 | WP_172694067 | TraY domain-containing protein | - |
| HTX04_RS00325 | 59023..59763 | - | 741 | WP_172694068 | hypothetical protein | - |
| HTX04_RS00330 | 59945..60409 | - | 465 | WP_247715244 | relaxosome protein TraM | - |
| HTX04_RS00335 | 60798..61247 | + | 450 | WP_172694069 | transglycosylase SLT domain-containing protein | virB1 |
| HTX04_RS00340 | 61491..62435 | - | 945 | WP_172694070 | zincin-like metallopeptidase domain-containing protein | - |
| HTX04_RS00345 | 62803..63477 | + | 675 | WP_172694071 | ParA family protein | - |
| HTX04_RS00350 | 63551..63844 | + | 294 | WP_036419818 | hypothetical protein | - |
| HTX04_RS00355 | 64006..64530 | + | 525 | WP_172694072 | SGNH/GDSL hydrolase family protein | - |
Host bacterium
| ID | 8239 | GenBank | NZ_MK851048 |
| Plasmid name | pMM1L5 | Incompatibility group | - |
| Plasmid size | 65134 bp | Coordinate of oriT [Strand] | 60636..60670 [+] |
| Host baterium | Morganella morganii strain MM1L5 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |