Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107662 |
| Name | oriT_OyaliB|p1 |
| Organism | Rhizobium leguminosarum strain OyaliB |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP071400 (95853..95881 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_OyaliB|p1
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file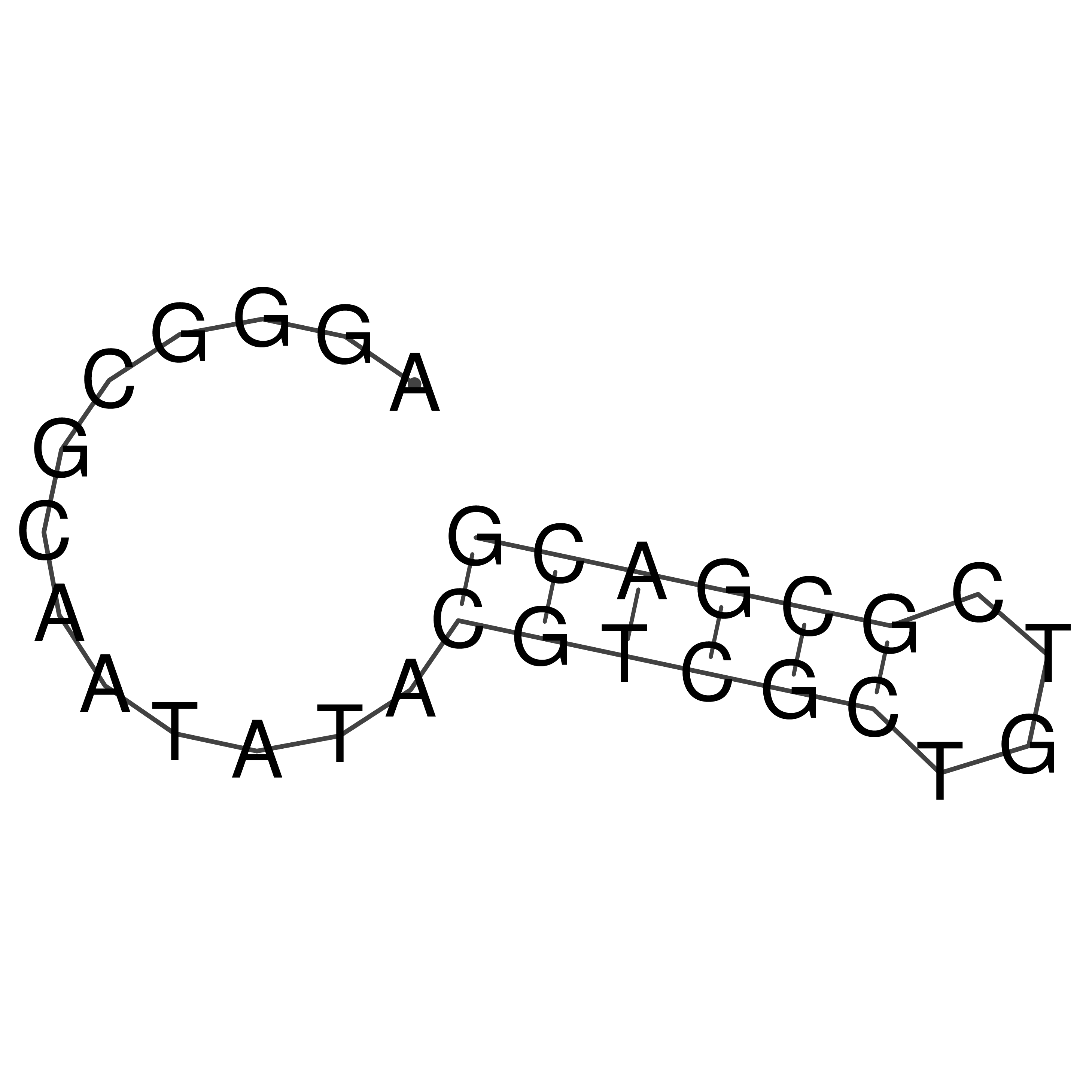
Relaxase
| ID | 5151 | GenBank | WP_207159192 |
| Name | traA_J0664_RS24360_OyaliB|p1 |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170671.04 Da Isoelectric Point: 7.5064
>WP_207159192.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAIMFVRAQVISRGVGRSIVSAAAYRHRARMMDEQAGTSFSYRGGRAELVHEELALPTQTPEWLRTAIDG
RTVAAASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTQEGFGRKKVAVTGEDGAPLRVVTPDRPKGRIVYKLWTGDKETMKAWKIAWAETANR
HLALAGHEIRIDGRSYAEQGLDGIAQRHLGPEKAALSRRGAELYFAPADLARRQEMADRLLADPALLLKQ
LGNERSTFDERDIAKALHRTVDDPLDFANIRARLMASDELVMLKPQQLDAETGKASEPAVFTTREILRIE
YDMARSAQVLSERCGFAVSDRVVAAAIRHVETQDLEKPFKLDAEQVDAVRHITGDSGIAAVVGLAGAGKS
TLLAAARVAWESGGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKLVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREEWAREASRLFARGE
IEKGLDAYAGHGHLVEAETRAETIDRIVADWTEARKQAIRNSTVVADDGRLRGDELLVLAHTNQDVKRLN
EALRSVMSGEGALGEGRSFQTERGAREFAAGDRIIFLENARFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GEVLLSVRLDNGRDVVLSEDSYRNVDHGYAATIHKSQGATVERTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEARPEWGRKPRVDHAAGVTGELVETGQAKFRPNDEDADDSPYADVKTDDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVEKVKVQVPIVDEQTGQKRFEEREVDRNVWTAKRIETADARLERIERESHRP
ELFKQLVLRLSRSGAKTTTLDFESEAGYQAHADDFARRRGIDTLSQAAAGMEEEVSRRWAWIAEKREQVA
ELWERASMALGFAIERERRVSYNEARIETQPVSDADAGRARHLIVPTTVFARTVDEDARLAQRSSPAWME
REAILRPLLEKIYRDPDAALVVLNALSSDASIEPRRLVDDLAATPDRLGRLRGSDLIVDGRAARDERSIA
TAALLELIPLARAHATEFRRNAERFGFREQQRRAHMSLPIPALSRQAMARLVEIEAVRDWGGDDAYKTAF
ALATEDRSLVQEVKAVSEALTARFGWSAFTSKADAIAERNIAGRLPEDLASTRREKLVGLFEAVKRFADE
QHLAERRDRSKIVAGASVERKNEHVAVLPMLAAVTEFKTPVDEEARSRVLSAPLYRQQRAVLAEAARAIW
RDPAGAVGKIEELLQKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVSEAAARLRA
LGSTYVNVLDGERQAIAEERRRMTVAIPGLSKAAEEALMRLTAEAKNNGRKLSAPAASLDPDIRREFEAV
SRALDERFGRDAILRQAKDRINRVPPPHRTAFEAMQDRLKILQQAVRMESSDRIISERRQRTANRGREI
MAIMFVRAQVISRGVGRSIVSAAAYRHRARMMDEQAGTSFSYRGGRAELVHEELALPTQTPEWLRTAIDG
RTVAAASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTQEGFGRKKVAVTGEDGAPLRVVTPDRPKGRIVYKLWTGDKETMKAWKIAWAETANR
HLALAGHEIRIDGRSYAEQGLDGIAQRHLGPEKAALSRRGAELYFAPADLARRQEMADRLLADPALLLKQ
LGNERSTFDERDIAKALHRTVDDPLDFANIRARLMASDELVMLKPQQLDAETGKASEPAVFTTREILRIE
YDMARSAQVLSERCGFAVSDRVVAAAIRHVETQDLEKPFKLDAEQVDAVRHITGDSGIAAVVGLAGAGKS
TLLAAARVAWESGGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKLVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREEWAREASRLFARGE
IEKGLDAYAGHGHLVEAETRAETIDRIVADWTEARKQAIRNSTVVADDGRLRGDELLVLAHTNQDVKRLN
EALRSVMSGEGALGEGRSFQTERGAREFAAGDRIIFLENARFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GEVLLSVRLDNGRDVVLSEDSYRNVDHGYAATIHKSQGATVERTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEARPEWGRKPRVDHAAGVTGELVETGQAKFRPNDEDADDSPYADVKTDDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVEKVKVQVPIVDEQTGQKRFEEREVDRNVWTAKRIETADARLERIERESHRP
ELFKQLVLRLSRSGAKTTTLDFESEAGYQAHADDFARRRGIDTLSQAAAGMEEEVSRRWAWIAEKREQVA
ELWERASMALGFAIERERRVSYNEARIETQPVSDADAGRARHLIVPTTVFARTVDEDARLAQRSSPAWME
REAILRPLLEKIYRDPDAALVVLNALSSDASIEPRRLVDDLAATPDRLGRLRGSDLIVDGRAARDERSIA
TAALLELIPLARAHATEFRRNAERFGFREQQRRAHMSLPIPALSRQAMARLVEIEAVRDWGGDDAYKTAF
ALATEDRSLVQEVKAVSEALTARFGWSAFTSKADAIAERNIAGRLPEDLASTRREKLVGLFEAVKRFADE
QHLAERRDRSKIVAGASVERKNEHVAVLPMLAAVTEFKTPVDEEARSRVLSAPLYRQQRAVLAEAARAIW
RDPAGAVGKIEELLQKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVSEAAARLRA
LGSTYVNVLDGERQAIAEERRRMTVAIPGLSKAAEEALMRLTAEAKNNGRKLSAPAASLDPDIRREFEAV
SRALDERFGRDAILRQAKDRINRVPPPHRTAFEAMQDRLKILQQAVRMESSDRIISERRQRTANRGREI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5391 | GenBank | WP_168342631 |
| Name | traG_J0664_RS24375_OyaliB|p1 |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70450.23 Da Isoelectric Point: 9.1439
>WP_168342631.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MALKCRLHPGLLLVLVPVAVTAMAVYITGWRWSALAHGLSGKSEYWFMRAAPVPALLFGPLTGLLTVWAL
PLHRRRSIAVASLLCFLVVSSFYALREYGRLAPSVESGTVSWDRALSYLDFVAVIGALAGFMAVAVAARI
SVVVPEQVKRAKGGTFGDADWLSMSAAAKLFPPDGEIVVGERYRVDKDIVHELPFDPNDRNTWGQGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYAGPLICLDPSTEVAPMVLEHRTRALGREVMVLDPT
NPIVGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSESAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNTFRSSDIIAGGTDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGRFSRRALFMLDEVDLLGYIRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFVSYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKRGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHRPIRCGRAIYFRRKDMNAAAK
ANRFVKPVP
MALKCRLHPGLLLVLVPVAVTAMAVYITGWRWSALAHGLSGKSEYWFMRAAPVPALLFGPLTGLLTVWAL
PLHRRRSIAVASLLCFLVVSSFYALREYGRLAPSVESGTVSWDRALSYLDFVAVIGALAGFMAVAVAARI
SVVVPEQVKRAKGGTFGDADWLSMSAAAKLFPPDGEIVVGERYRVDKDIVHELPFDPNDRNTWGQGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYAGPLICLDPSTEVAPMVLEHRTRALGREVMVLDPT
NPIVGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSESAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNTFRSSDIIAGGTDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGRFSRRALFMLDEVDLLGYIRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFVSYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKRGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHRPIRCGRAIYFRRKDMNAAAK
ANRFVKPVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 650881..660488
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| J0664_RS36305 | 646799..646984 | + | 186 | WP_246767367 | hypothetical protein | - |
| J0664_RS36310 | 646971..647168 | + | 198 | WP_246767368 | hypothetical protein | - |
| J0664_RS26800 (J0664_26800) | 647477..648086 | + | 610 | Protein_585 | integrase | - |
| J0664_RS26805 (J0664_26805) | 648161..648595 | - | 435 | WP_207158858 | type II toxin-antitoxin system VapC family toxin | - |
| J0664_RS26810 (J0664_26810) | 648595..648837 | - | 243 | WP_046601781 | type II toxin-antitoxin system Phd/YefM family antitoxin | - |
| J0664_RS26815 (J0664_26815) | 649271..650179 | + | 909 | WP_207158859 | nucleotidyltransferase and HEPN domain-containing protein | - |
| J0664_RS26820 (J0664_26820) | 650510..650725 | + | 216 | WP_207158860 | DUF3072 domain-containing protein | - |
| J0664_RS26825 (J0664_26825) | 650881..651921 | - | 1041 | WP_207158861 | P-type DNA transfer ATPase VirB11 | virB11 |
| J0664_RS26830 (J0664_26830) | 651929..653101 | - | 1173 | WP_207158862 | type IV secretion system protein VirB10 | virB10 |
| J0664_RS26835 (J0664_26835) | 653111..653968 | - | 858 | WP_207158863 | P-type conjugative transfer protein VirB9 | virB9 |
| J0664_RS26840 (J0664_26840) | 653965..654636 | - | 672 | WP_207158864 | virB8 family protein | virB8 |
| J0664_RS26845 (J0664_26845) | 654638..654922 | - | 285 | WP_207158865 | hypothetical protein | - |
| J0664_RS26850 (J0664_26850) | 654974..655894 | - | 921 | WP_207158866 | type IV secretion system protein | virB6 |
| J0664_RS26855 (J0664_26855) | 655898..656131 | - | 234 | WP_207158867 | EexN family lipoprotein | - |
| J0664_RS26860 (J0664_26860) | 656128..656565 | - | 438 | WP_348651639 | P-type DNA transfer protein VirB5 | - |
| J0664_RS36695 | 656474..656827 | - | 354 | WP_348651640 | type IV secretion system protein | - |
| J0664_RS26865 (J0664_26865) | 656827..659193 | - | 2367 | WP_207158868 | VirB4 family type IV secretion system protein | virb4 |
| J0664_RS26870 (J0664_26870) | 659186..659524 | - | 339 | WP_062941069 | type IV secretion system protein VirB3 | virB3 |
| J0664_RS26875 (J0664_26875) | 659530..659829 | - | 300 | WP_065282894 | TrbC/VirB2 family protein | virB2 |
| J0664_RS26880 (J0664_26880) | 659826..660488 | - | 663 | WP_207158869 | lytic transglycosylase domain-containing protein | virB1 |
| J0664_RS26885 (J0664_26885) | 660492..661031 | - | 540 | WP_207158870 | hypothetical protein | - |
| J0664_RS26890 (J0664_26890) | 661123..661476 | + | 354 | WP_168342759 | MarR family transcriptional regulator | - |
| J0664_RS26895 (J0664_26895) | 662142..663137 | + | 996 | WP_207159451 | glycosyltransferase | - |
Host bacterium
| ID | 8097 | GenBank | NZ_CP071400 |
| Plasmid name | OyaliB|p1 | Incompatibility group | - |
| Plasmid size | 1230906 bp | Coordinate of oriT [Strand] | 95853..95881 [-] |
| Host baterium | Rhizobium leguminosarum strain OyaliB |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | AcrIIA7 |