Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107619 |
| Name | oriT_pKJK172 |
| Organism | Thauera aromatica K172 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP028340 (44228..44302 [+], 75 nt) |
| oriT length | 75 nt |
| IRs (inverted repeats) | 25..32, 35..42 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 61..62 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 75 nt
CGAATAAGGGACAGTGAAGATAGATAACCGGCTTGCCGGTTAGCTAACTTCACCCATCCTGCCCGCCTTACGGCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file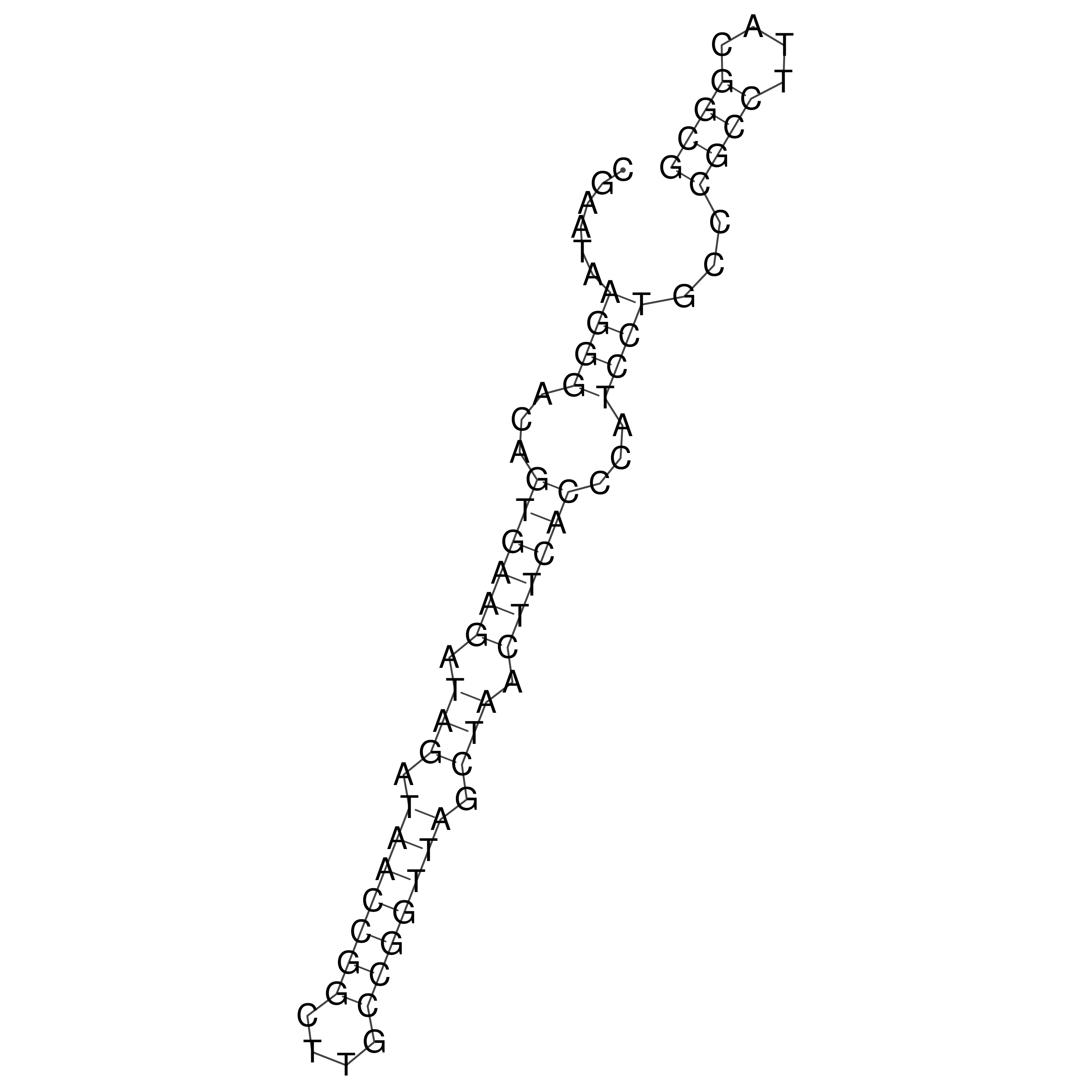
Relaxase
| ID | 5121 | GenBank | WP_011600636 |
| Name | Relaxase_Tharo_RS17430_pKJK172 |
UniProt ID | A0A899NDK5 |
| Length | 740 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 740 a.a. Molecular weight: 81453.36 Da Isoelectric Point: 10.7289
MIAKHVPMRSLGKSDFAGLVQYITDAQSKTERLGLVTLTNCQAGTVQAATDEVLATQHMNTRATGDKTFH
LLVSFRAGEKPDADTLKAIEERICAGLGYGEHQRISAVHHDTDNLHIHIAINKIHPTRNTMHEPFQSYRT
LGELCEVLERDYGLEKDNHTPARSIAEGRAADMERHAGVESLVGWIKRECLEEIKAAQSWTELHQALQAN
GLELRAKGNGFVIEAGDGTQVKASTVARELSKPKLEARFGPFEASPERQAQTKAKRQYQKKPVRLRVNTT
ELYARYKADQQTLAAAQRAALDKARQSKTRKVEDAKRSSRLRRAAIKLMGGKAATKKLLYAQASKALRDE
IQAINKEHQRERQGCYDEHKRRTWADWLKKEALQGNAEALAALRAREAAQGLKGNTVEGQGQAKPGHAPV
VDNITKKGTIIFRAGKTAVRDDGDKLQVSREATREGLQQALRLAMERYGSRITVNGTTEFKAQIIRAAVD
SQLPITFADPALESRRQALLKKENTHDRPQQDRGRTGRGTGRPGSRPAADNDAARSSVNADRGGRDPAGR
VHATGQHRKPDIGRIGRVPPPQSKNRLRALSKLGVVRIASGSEVLLPRDVPGHMEQQGTQPDNALRRGVS
RPGGRLEAGPQYKPEQVKAMDKYIAEREGKRLKGFDIPKHSRYTAGDGALSFAGMRNVEGQTLALVKRGD
DVLVMPIDQATANRLKRIAVGDAVSITPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A899NDK5 |
Auxiliary protein
| ID | 2342 | GenBank | WP_011600638 |
| Name | WP_011600638_pKJK172 |
UniProt ID | A0A899NCL8 |
| Length | 124 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 124 a.a. Molecular weight: 14196.30 Da Isoelectric Point: 10.1233
MEKDEKQTGRKRRHHLRVPVFPDEKEEIERQARQAGLSVARYLREVGQGYEIKGITDYERVRELARINGD
LGRLGGLLKLWLTDDARTAKFGEATILALLGRIEATQDEMGRVMKAVVRPRAEP
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A899NCL8 |
T4CP
| ID | 5352 | GenBank | WP_011600613 |
| Name | t4cp2_Tharo_RS17220_pKJK172 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 94237.23 Da Isoelectric Point: 6.2155
MIETIAFAIAVLGAVLLLILFARIRAVDAELKLKKHRSKDAGLADLLNYAAVVDDGVIVCKNGSFMASWL
YKGDDNASSTDEQREMVSFRINQALAGLGSGWMVHVDAVRRPAPNYSDRGHSNFPDPLSAAIDEERRQLF
QGLGTMYEGYFVLTLTWFPPLLAQRKFVELMFDDDAVVPDHKARTMGLIKQFKREITSIESRLSSAVKLT
RLRGQKVVTEAGREVTHDDFLAWLQFCITGLHHPVQLPSNPMYLDALIGGQEMFGGVVPKVGRNFVQVVA
VEGFPLESTPGVLTALGELACEYRWSSRFIFMDQHEAVAHLDKFRKKWRQKVRGFFDQVFNTNSGNIDQD
ALSMVGDAEAAIAEVNSGLVAVGYYTGVVVLMDEDRTRLEQSARQVEKAINRLGFAARIETINTLDAFLG
SLPGHGVENVRRPLINTMNLADLLPTSTIWTGLASAPCPMYPPLSPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYTDMSIFAFDKGMSMYPLAAGIRAGTKGKSGLHFTVAADDDQLAFC
PLQFLETKGDRAWAMEWIDTILALNGVETTPAQRNAISNAIFSMHDSGAHTLSEFSVTIQDEAIREAIQQ
YTVDGTMGHLLDAETDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALHGQPAVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVAAIKNLEAKFGDAWVHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5353 | GenBank | WP_011600635 |
| Name | t4cp2_Tharo_RS17425_pKJK172 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69544.72 Da Isoelectric Point: 8.2324
MKIKMNNAVGPQVRSAKPKAGKLLPTLGVLSLAGGLQAATQYFAHTFDYQASLGGHIGHIYAPWSILQWA
SKWYSHYPNEIMSAGSVGMLVSTVGLLGVAIAKVVASNTSKANQFLHGSARWAEKKDIQAAGLLPRPRTV
LEVVTGKEEPTATGVYVGGWEDKDGSFYYLRHSGPEHVLCYAPTRSGKGVGLVVPTMLSWPASAVITDLK
GELWALTAGWRKKHAKNKVLRFEPASSNGGICWNPLDEIRLGTEFEVGDVQNLAQLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAQADGTYATLPSVDAMLADPNRDVGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLSLYRDPVVARSVSKSEFRIKDLMHQDDPASLYIVTQPNDKARLRPLVRIMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDETITSNCHVQNAYPPNRVETAEHLSRLTGQTTIVKEQITTSGRRTAAMLGQVSRTFQEVQRPLLT
PDECLRMPGPRKSATGEIEEAGDMVVYVAGYPAIYGKQPLYFKDPVFQARAAIPAPKVSDKTIHVQDAGQ
GITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2928..13896
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Tharo_RS17190 (Tharo_3405) | 536..1759 | - | 1224 | WP_011600607 | plasmid replication initiator TrfA | - |
| Tharo_RS17195 (Tharo_3406) | 1811..2152 | - | 342 | WP_011600608 | single-stranded DNA-binding protein | - |
| Tharo_RS17200 (Tharo_3407) | 2264..2638 | + | 375 | WP_011600609 | transcriptional regulator | - |
| Tharo_RS17205 (Tharo_3409) | 2928..3890 | + | 963 | WP_011600610 | P-type conjugative transfer ATPase TrbB | virB11 |
| Tharo_RS17210 (Tharo_3410) | 3904..4368 | + | 465 | WP_011600611 | conjugal transfer system pilin TrbC | virB2 |
| Tharo_RS17215 (Tharo_3411) | 4372..4683 | + | 312 | WP_011600612 | conjugal transfer protein TrbD | virB3 |
| Tharo_RS17220 (Tharo_3412) | 4680..7238 | + | 2559 | WP_011600613 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| Tharo_RS17225 (Tharo_3413) | 7235..8008 | + | 774 | WP_011600614 | conjugal transfer protein TrbF | virB8 |
| Tharo_RS17230 (Tharo_3414) | 8026..8919 | + | 894 | WP_011600615 | P-type conjugative transfer protein TrbG | virB9 |
| Tharo_RS17235 (Tharo_3415) | 8922..9377 | + | 456 | WP_011600616 | lipoprotein | - |
| Tharo_RS17240 (Tharo_3416) | 9381..10784 | + | 1404 | WP_011600617 | TrbI/VirB10 family protein | virB10 |
| Tharo_RS17245 (Tharo_3417) | 10803..11573 | + | 771 | WP_011600618 | P-type conjugative transfer protein TrbJ | virB5 |
| Tharo_RS17250 (Tharo_3418) | 11583..11756 | + | 174 | WP_011600619 | entry exclusion lipoprotein TrbK | - |
| Tharo_RS17255 (Tharo_3419) | 11753..11968 | + | 216 | WP_031943485 | entry exclusion lipoprotein TrbK | - |
| Tharo_RS17260 (Tharo_3420) | 11980..13896 | + | 1917 | WP_011600621 | P-type conjugative transfer protein TrbL | virB6 |
| Tharo_RS17265 (Tharo_3421) | 13932..14519 | + | 588 | WP_011600622 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| Tharo_RS17270 (Tharo_3422) | 14539..15165 | + | 627 | WP_042993594 | transglycosylase SLT domain-containing protein | - |
| Tharo_RS17275 (Tharo_3423) | 15194..15463 | + | 270 | WP_011600624 | hypothetical protein | - |
| Tharo_RS17280 (Tharo_3424) | 15460..16158 | + | 699 | WP_011600625 | TraX family protein | - |
| Tharo_RS17285 (Tharo_3425) | 16173..16604 | + | 432 | WP_011600626 | OmpA family protein | - |
| Tharo_RS17290 (Tharo_3426) | 16647..17060 | + | 414 | WP_011600627 | hypothetical protein | - |
| Tharo_RS17295 (Tharo_3427) | 17086..18054 | + | 969 | WP_011600628 | hypothetical protein | - |
| Tharo_RS17300 | 18037..18642 | - | 606 | WP_011600629 | recombinase family protein | - |
Host bacterium
| ID | 8054 | GenBank | NZ_CP028340 |
| Plasmid name | pKJK172 | Incompatibility group | - |
| Plasmid size | 53761 bp | Coordinate of oriT [Strand] | 44228..44302 [+] |
| Host baterium | Thauera aromatica K172 |
Cargo genes
| Drug resistance gene | ere(A), qacE, sul1, tet(A) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |