Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107607 |
| Name | oriT_pSF45436d |
| Organism | Sinorhizobium fredii CCBAU 45436 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP029234 (34746..34774 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSF45436d
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file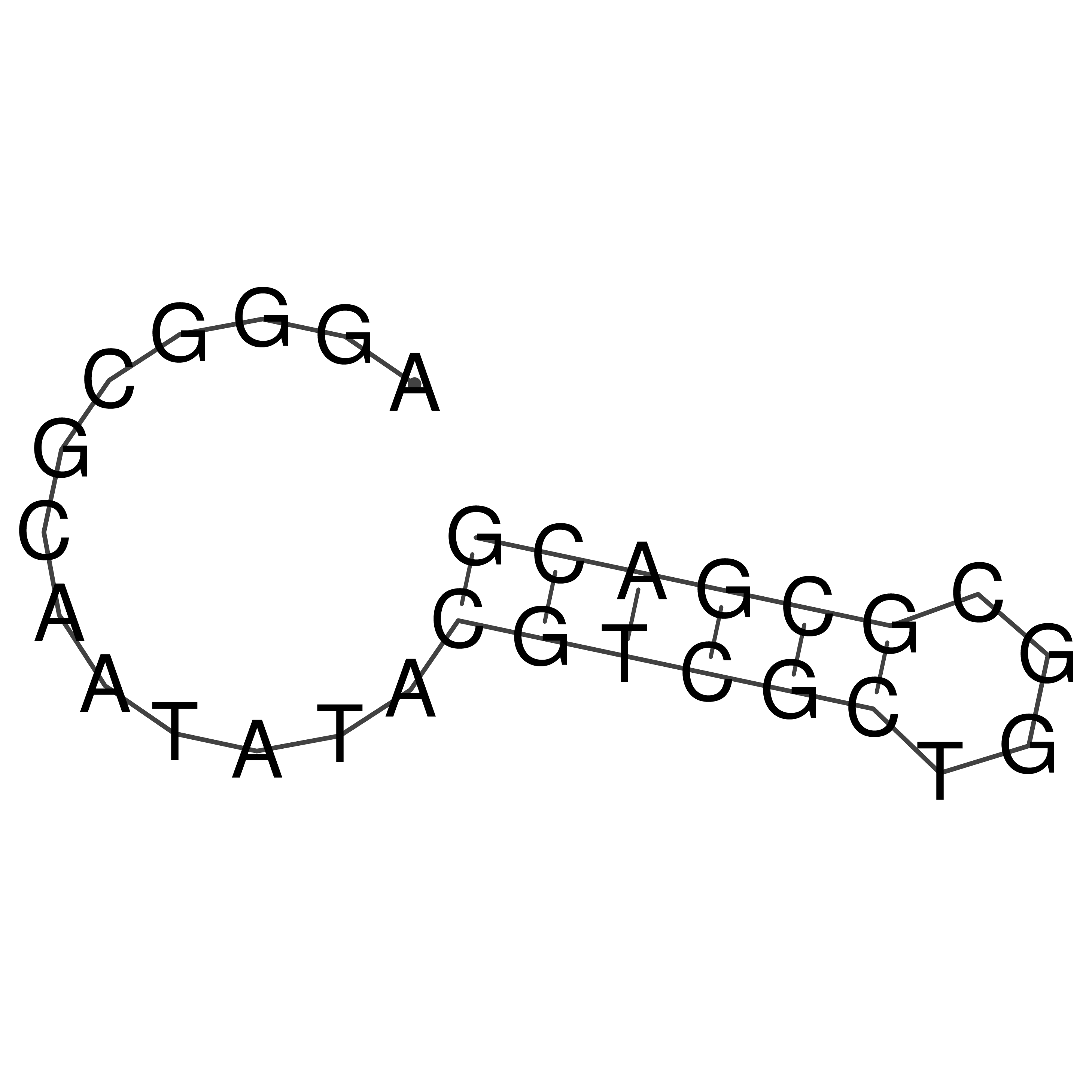
Relaxase
| ID | 5114 | GenBank | WP_015633563 |
| Name | traA_AB395_RS32180_pSF45436d |
UniProt ID | A0A844A347 |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170749.69 Da Isoelectric Point: 8.8055
>WP_015633563.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Sinorhizobium]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPDQIPGWLKAAIDR
QSVAKASEALWNAVESHETRADAQLARELIIALPEELTRAENIALVREFIRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEDGEPLRVVTPDRPNGKIVYQLWAGDKETMKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLHGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLIADPELLLKQ
LGSERSTFEERDIAKALHRYVDDPVDFVNIRARLMASDDLVLLKPQEVDAQTGRASEPAVFTTREILRIE
YDMAQSAQALSERKGFGVSTKQVAAAIRNVETADPEEPFKLHAEQVDAVRHVTGDSGMAAVVGLAGAGKS
TLLAAARVAWESEDRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWAGGREQLDRGDVLVIDEAGMIS
SQQMARVLKIAQEAQAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREQWARDASRLFARGK
VEEGLDAYAQQGHIVETETRAEIVDRIVSDWTDARRDLLQKSAAGEHPGRLRGEELLVLAHTNQDVRKLN
ESLRKVMIDEGALTAAREFQTARGSREFAAGDRIIFLENARFIEPRARRLGPQYVKNGMLGTVVSTGDKR
GCALLTVRLDNGRDVVISEDSYRNIDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRHRADL
YVAKEDFEVRPEWGRKPRVDHAAGVIGELVETGEAKFRPQERQAKESPYADVRTDDGTVHRLWGVSLPKA
LEEGSVSTGDTVTLRKDGVEKVIVKIPVTDEETGEKRLEEREVERNVWTANRVETADARQERIKRERHRL
ELFKQLVERLGRSGAKTTTLDFASEEVYQAHARDFARRRGIDTLAEVAAGMEEGVSRRLAWIAEKRGQVA
KLWERASVALGFAIERERRVSYNEERTASFSAGIPLVGKYLMPPTTKFSRSVEEDARLAQLSSERWKERE
AVLRPVLAKIYLDPDGALSALNALASDTAIEPRKLAENLGSVPDRLGRLRGSNLMVDGRAARDDRTAATA
AVSELLPLARAHSTEFRRQAERFGIREQQRRAHMSLSVPALSKTAMARLVEIEAVCDRGGADAYKTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVVAERNMFERMPEDIPPERREKLARLFAVIRRFVEEQH
LAERQDRSKIVAGASVEWGKETMAVLPMLPAVTEFKTTVADEAKERALAAPHYGHHRAALVETATRVWRD
PADAIGNIEELVVKGFASERIAAAVSNDPAAYGALRGSDRIMDKLLTTGRERKEALQAVPEVASRVRSLG
ASYASAVDAETQAIAEERRRMAIAIPGLSQAAEDALKRLAAEIKKKDGKLDVAAGSLDARIRQEFAGVSR
ALDERFGRNAILRGEKDVINRVSPAQRRAFEAMRDRLQVLQQTVRLQSSQEIISERQRRVIDRARGVTR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPDQIPGWLKAAIDR
QSVAKASEALWNAVESHETRADAQLARELIIALPEELTRAENIALVREFIRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEDGEPLRVVTPDRPNGKIVYQLWAGDKETMKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLHGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLIADPELLLKQ
LGSERSTFEERDIAKALHRYVDDPVDFVNIRARLMASDDLVLLKPQEVDAQTGRASEPAVFTTREILRIE
YDMAQSAQALSERKGFGVSTKQVAAAIRNVETADPEEPFKLHAEQVDAVRHVTGDSGMAAVVGLAGAGKS
TLLAAARVAWESEDRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWAGGREQLDRGDVLVIDEAGMIS
SQQMARVLKIAQEAQAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREQWARDASRLFARGK
VEEGLDAYAQQGHIVETETRAEIVDRIVSDWTDARRDLLQKSAAGEHPGRLRGEELLVLAHTNQDVRKLN
ESLRKVMIDEGALTAAREFQTARGSREFAAGDRIIFLENARFIEPRARRLGPQYVKNGMLGTVVSTGDKR
GCALLTVRLDNGRDVVISEDSYRNIDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRHRADL
YVAKEDFEVRPEWGRKPRVDHAAGVIGELVETGEAKFRPQERQAKESPYADVRTDDGTVHRLWGVSLPKA
LEEGSVSTGDTVTLRKDGVEKVIVKIPVTDEETGEKRLEEREVERNVWTANRVETADARQERIKRERHRL
ELFKQLVERLGRSGAKTTTLDFASEEVYQAHARDFARRRGIDTLAEVAAGMEEGVSRRLAWIAEKRGQVA
KLWERASVALGFAIERERRVSYNEERTASFSAGIPLVGKYLMPPTTKFSRSVEEDARLAQLSSERWKERE
AVLRPVLAKIYLDPDGALSALNALASDTAIEPRKLAENLGSVPDRLGRLRGSNLMVDGRAARDDRTAATA
AVSELLPLARAHSTEFRRQAERFGIREQQRRAHMSLSVPALSKTAMARLVEIEAVCDRGGADAYKTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVVAERNMFERMPEDIPPERREKLARLFAVIRRFVEEQH
LAERQDRSKIVAGASVEWGKETMAVLPMLPAVTEFKTTVADEAKERALAAPHYGHHRAALVETATRVWRD
PADAIGNIEELVVKGFASERIAAAVSNDPAAYGALRGSDRIMDKLLTTGRERKEALQAVPEVASRVRSLG
ASYASAVDAETQAIAEERRRMAIAIPGLSQAAEDALKRLAAEIKKKDGKLDVAAGSLDARIRQEFAGVSR
ALDERFGRNAILRGEKDVINRVSPAQRRAFEAMRDRLQVLQQTVRLQSSQEIISERQRRVIDRARGVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5340 | GenBank | WP_014857689 |
| Name | traG_AB395_RS32165_pSF45436d |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70288.20 Da Isoelectric Point: 9.2510
>WP_014857689.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Sinorhizobium]
MALKAKPHPSLLLVLVPVAVTAFAVYVVGWRWQALAEGMSGKTQYWFLRVSPVPALLFGPLAGLLAVWAL
PLHRRRPVAMAGLACFLTVAGFYAVREFGRLAPSVESGALSWDRALSYLDMVAVVGAVVGFIAVAIAARI
STVVSEPVKRAKLGTFGDADWLPMSAAGKLFPPDGEIVVGERYRVDKDIVHELPFDPNDPAAWGKGGKTP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESIRFESSTGSYFQNQAHNLLTGLLTHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGNKDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGGFKRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKPIP
MALKAKPHPSLLLVLVPVAVTAFAVYVVGWRWQALAEGMSGKTQYWFLRVSPVPALLFGPLAGLLAVWAL
PLHRRRPVAMAGLACFLTVAGFYAVREFGRLAPSVESGALSWDRALSYLDMVAVVGAVVGFIAVAIAARI
STVVSEPVKRAKLGTFGDADWLPMSAAGKLFPPDGEIVVGERYRVDKDIVHELPFDPNDPAAWGKGGKTP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESIRFESSTGSYFQNQAHNLLTGLLTHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGNKDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGGFKRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKPIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 189211..198788
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AB395_RS32860 (AB395_00006725) | 185698..186354 | - | 657 | WP_014857948 | CBS domain-containing protein | - |
| AB395_RS32865 (AB395_00006727) | 186690..186884 | + | 195 | WP_014857951 | hypothetical protein | - |
| AB395_RS32870 (AB395_00006728) | 186893..188491 | + | 1599 | WP_014857952 | AarF/UbiB family protein | - |
| AB395_RS35150 | 188554..188730 | + | 177 | Protein_177 | methionine adenosyltransferase | - |
| AB395_RS32880 (AB395_00006730) | 189211..190200 | - | 990 | WP_014857954 | P-type DNA transfer ATPase VirB11 | virB11 |
| AB395_RS32885 (AB395_00006731) | 190211..191383 | - | 1173 | WP_037457198 | type IV secretion system protein VirB10 | virB10 |
| AB395_RS32890 (AB395_00006732) | 191393..192247 | - | 855 | WP_037457195 | P-type conjugative transfer protein VirB9 | virB9 |
| AB395_RS32895 (AB395_00006733) | 192247..192918 | - | 672 | WP_014857957 | virB8 family protein | virB8 |
| AB395_RS32900 (AB395_00006734) | 192923..193198 | - | 276 | WP_014857958 | hypothetical protein | - |
| AB395_RS32905 (AB395_00006735) | 193235..194170 | - | 936 | WP_037457191 | type IV secretion system protein | virB6 |
| AB395_RS32910 (AB395_00006736) | 194176..194406 | - | 231 | WP_014857960 | EexN family lipoprotein | - |
| AB395_RS32915 (AB395_00006737) | 194403..195101 | - | 699 | WP_037457188 | P-type DNA transfer protein VirB5 | virB5 |
| AB395_RS32920 (AB395_00006738) | 195101..197482 | - | 2382 | WP_014857962 | VirB4 family type IV secretion system protein | virb4 |
| AB395_RS32925 (AB395_00006739) | 197475..197816 | - | 342 | WP_014857963 | type IV secretion system protein VirB3 | virB3 |
| AB395_RS32930 (AB395_00006740) | 197821..198120 | - | 300 | WP_014857964 | TrbC/VirB2 family protein | virB2 |
| AB395_RS32935 (AB395_00006741) | 198117..198788 | - | 672 | WP_014857965 | lytic transglycosylase domain-containing protein | virB1 |
| AB395_RS32940 (AB395_00006742) | 198792..199310 | - | 519 | WP_014857966 | hypothetical protein | - |
| AB395_RS32945 (AB395_00006743) | 199407..199775 | + | 369 | WP_014857967 | transcriptional regulator | - |
| AB395_RS32950 | 199994..200672 | - | 679 | Protein_192 | acyl-homoserine-lactone synthase TraI | - |
Host bacterium
| ID | 8042 | GenBank | NZ_CP029234 |
| Plasmid name | pSF45436d | Incompatibility group | - |
| Plasmid size | 201059 bp | Coordinate of oriT [Strand] | 34746..34774 [+] |
| Host baterium | Sinorhizobium fredii CCBAU 45436 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |