Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107603 |
| Name | oriT_pRLN3 |
| Organism | Rhizobium leguminosarum strain Norway |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP025015 (537136..537169 [+], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file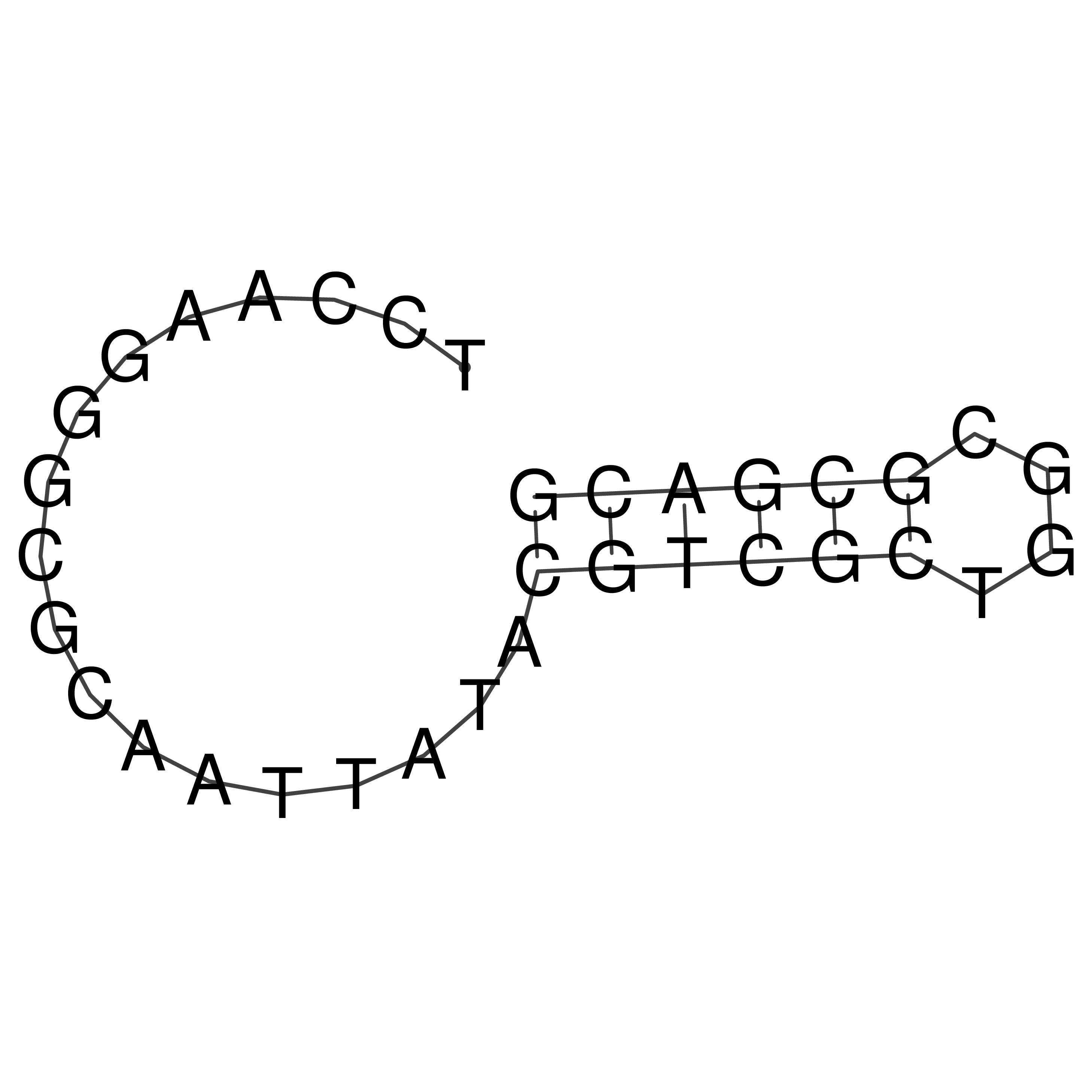
Relaxase
| ID | 5112 | GenBank | WP_105009996 |
| Name | traA_CUJ84_RS34840_pRLN3 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122765.90 Da Isoelectric Point: 9.7558
MAVPHFSVSVVARGSGRSAVLSAAYRHCTKMEFEREARTIDYTRKQGLLHEEFIIPADAPAWLRAMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVPEAVLQATFARHSRLSDEQRTAIEHIAGTTRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDRCVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMLGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLAYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYE
KSALYRQALRFAEARGLHLMNVARTIAHDRLQWAVRQSSRLADLGARLAAIGKALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPEAAFKAINVDAMLKDQSVAQS
TLARIAGEPESFGALKGNTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPSAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDAGG
KTFETVTTGMTPGQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5335 | GenBank | WP_105009699 |
| Name | t4cp2_CUJ84_RS32525_pRLN3 |
UniProt ID | _ |
| Length | 577 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 577 a.a. Molecular weight: 64544.32 Da Isoelectric Point: 9.3996
MSNKFDLFGDLERHLSLALRWGKWGAKTGRNLKARSARQHVEELYGPEVADAVDAAVRSGRDPGPIIERA
KRDLAARQERERQIADPPPLYGSARWPTSQDLRPYLRDRKAFDDPRSIQLGTFSDEGQATPAGFAHWDGD
GHLLTIAPTRSGKGKTTIIPNLLRYQGSCVVLDPKGELFEATSQWRSTLGPVYRIAPLDGGSGKPVHRFD
PVSRVRRESDARAIAMQMFPRDPKSPAFFADDAVGFMTAIVMFVRYKAPPHRRTLATVCQMASLKGASLL
KVAKEMEAFPPSADGARASLEKDPLRGLKVLQETLTSKLYKWKDADIQRSVSGNDVSFESLKDRPATVYI
EIPFDMMKTFSGWLRVVLKTALDAMLTNPRVPDIPVLFVLDEFLNIGAFPEYRDAIRTHAGAGIRLWFFL
QDIYSIEEHYPGNSWRPFLNCAVKQFFGINDDDTAKMIGGYLGHKTHAIRTTRASASISSHLGEWYGDAS
TNTSFSMTESVQFLGRPLLTSDEVRELLGGWQGDGWRYGITDIAGTRPFKTQLVVHEKSETCRQRIGLMT
RGESDDRNDHQLSAPQK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5336 | GenBank | WP_105009994 |
| Name | traG_CUJ84_RS34825_pRLN3 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69482.93 Da Isoelectric Point: 9.6636
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRARIALPYAASAAIGVLFLFACAGSI
NIKRAGWGVLAGCIGTMLIAAVRETLRLSALMKTLPAGKTVLAYLDPATSIGASAALLCAGFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDIVGSQAFRADSAENWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRRVRMNLSEPEPTLRKRLQDVYDNSGSDFVKENVAAFVNLTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQLNGRALFLLDEVARLGY
MRILETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETAEYISKRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIIFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5337 | GenBank | WP_018482025 |
| Name | t4cp2_CUJ84_RS34925_pRLN3 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91596.40 Da Isoelectric Point: 5.9999
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVESRFLGVVAIDGLPVESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARRKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIARASSQLVAYGYYTPVVVL
FGSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDSVEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDSLHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVQDAESLLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 546875..556375
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CUJ84_RS34855 (CUJ84_pRLN3000586) | 542306..542923 | + | 618 | WP_105009999 | TraH family protein | - |
| CUJ84_RS34860 (CUJ84_pRLN3000588) | 543054..543920 | - | 867 | WP_105010000 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| CUJ84_RS34865 (CUJ84_pRLN3000589) | 543913..544530 | - | 618 | WP_105010001 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| CUJ84_RS34870 (CUJ84_pRLN3000590) | 544790..545062 | - | 273 | Protein_488 | DUF433 domain-containing protein | - |
| CUJ84_RS34875 (CUJ84_pRLN3000591) | 545114..545326 | - | 213 | WP_018482035 | helix-turn-helix transcriptional regulator | - |
| CUJ84_RS34880 (CUJ84_pRLN3000592) | 545532..545855 | + | 324 | WP_018482034 | transcriptional repressor TraM | - |
| CUJ84_RS34885 (CUJ84_pRLN3000593) | 545859..546563 | - | 705 | WP_105010002 | autoinducer binding domain-containing protein | - |
| CUJ84_RS34890 (CUJ84_pRLN3000594) | 546875..548173 | - | 1299 | WP_018482032 | IncP-type conjugal transfer protein TrbI | virB10 |
| CUJ84_RS34895 (CUJ84_pRLN3000595) | 548175..548618 | - | 444 | WP_018482031 | conjugal transfer protein TrbH | - |
| CUJ84_RS34900 (CUJ84_pRLN3000596) | 548622..549434 | - | 813 | WP_020572024 | P-type conjugative transfer protein TrbG | virB9 |
| CUJ84_RS34905 (CUJ84_pRLN3000597) | 549452..550114 | - | 663 | WP_018482029 | conjugal transfer protein TrbF | virB8 |
| CUJ84_RS34910 (CUJ84_pRLN3000598) | 550135..551313 | - | 1179 | WP_105010003 | P-type conjugative transfer protein TrbL | virB6 |
| CUJ84_RS34915 (CUJ84_pRLN3000599) | 551327..551503 | - | 177 | WP_018482027 | entry exclusion protein TrbK | - |
| CUJ84_RS34920 (CUJ84_pRLN3000600) | 551497..552294 | - | 798 | WP_018482026 | P-type conjugative transfer protein TrbJ | virB5 |
| CUJ84_RS34925 (CUJ84_pRLN3000601) | 552272..554722 | - | 2451 | WP_018482025 | conjugal transfer protein TrbE | virb4 |
| CUJ84_RS34930 (CUJ84_pRLN3000602) | 554733..555032 | - | 300 | WP_018482024 | conjugal transfer protein TrbD | virB3 |
| CUJ84_RS34935 (CUJ84_pRLN3000603) | 555025..555408 | - | 384 | WP_018482023 | TrbC/VirB2 family protein | virB2 |
| CUJ84_RS34940 (CUJ84_pRLN3000604) | 555398..556375 | - | 978 | WP_018482022 | P-type conjugative transfer ATPase TrbB | virB11 |
| CUJ84_RS34945 (CUJ84_pRLN3000605) | 556372..557010 | - | 639 | WP_018482021 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 8038 | GenBank | NZ_CP025015 |
| Plasmid name | pRLN3 | Incompatibility group | - |
| Plasmid size | 557386 bp | Coordinate of oriT [Strand] | 537136..537169 [+] |
| Host baterium | Rhizobium leguminosarum strain Norway |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | hupE |
| Degradation gene | - |
| Symbiosis gene | fixQ, fixO, fixN, nifN, nifE, nifK, nifD, nifH, fixA, fixB, fixC, fixX, nifB, nodJ, nodI, nodC, nodA, nodD, nodF, nodE, nodM |
| Anti-CRISPR | - |