Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107597 |
| Name | oriT_pSK02 |
| Organism | Rhizobium leguminosarum bv. viciae strain BIHB 1148 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP022566 (34724..34752 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSK02
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file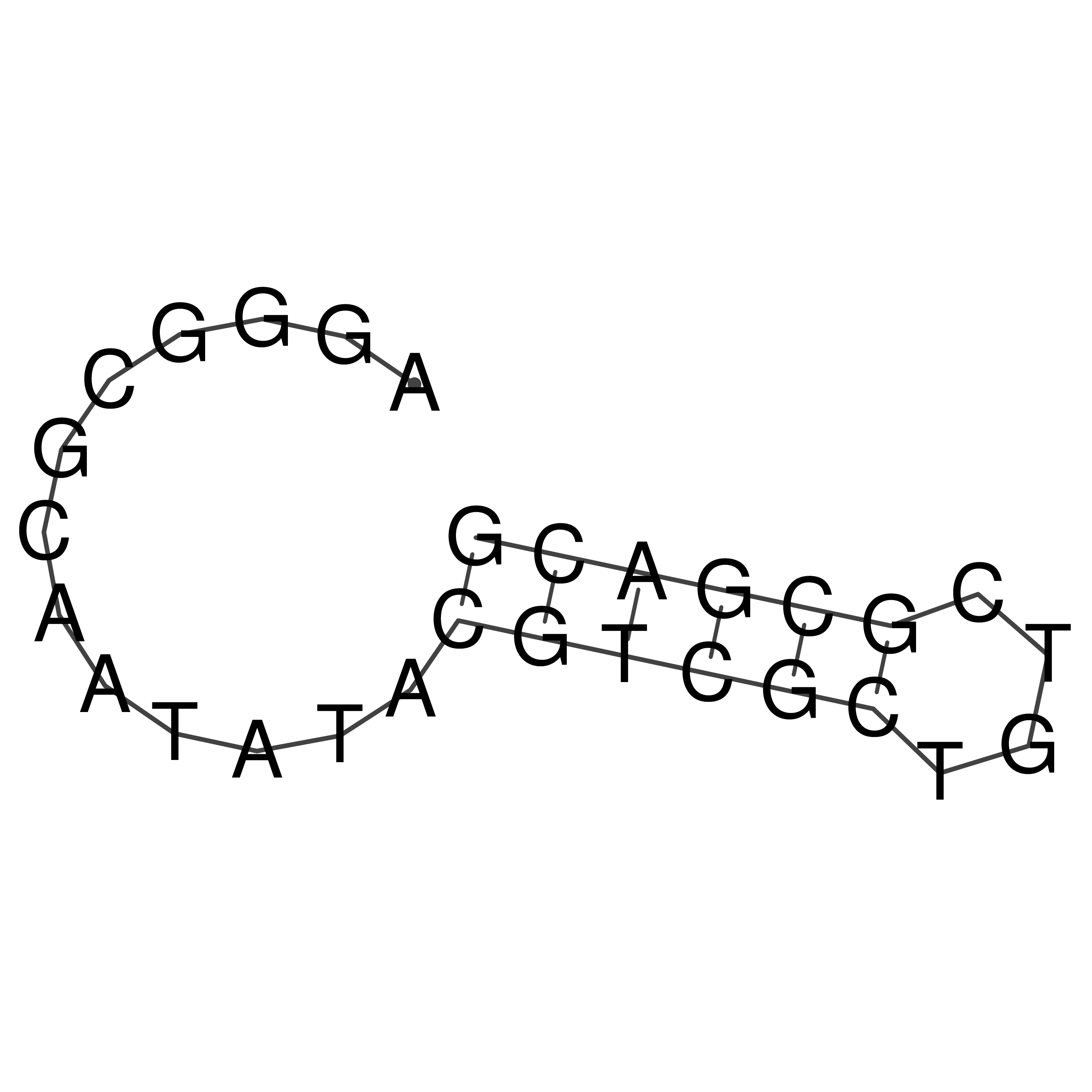
Relaxase
| ID | 5107 | GenBank | WP_094088384 |
| Name | traA_CHY08_RS26900_pSK02 |
UniProt ID | _ |
| Length | 1545 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1545 a.a. Molecular weight: 171110.24 Da Isoelectric Point: 6.6888
>WP_094088384.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAIMFVRAQVISRGVGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAPELVHEEVALPDWTPAWLRAAIDG
RTVAAASEALWNAVDVFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTQEGFGRKKVAVTGEDGAPLRVVTPDRPKGRIVYKLWAGDKETLQAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALSRRGAEMYFAPADLARRQEMADRLLADPTLLLKQ
LGNERSTFDERDIAKALHRYVDDPADFANIRARLMASDDLVMLKPQQVDAETGKASEPAVFTTREILRVE
YDMSQSARVLSERRGFAVSAESVAAAIENVETADPEKSFRLDAEQVDAVRHITGDSGIAAVVGLAGAGKS
TLLAAARLAWEHDGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELIWADGHETLHRGDVIVIDEAGMVS
SQQMARVLKLVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREDWAREASRLFARGE
IEKGLDAYAQQGHLIEAATRDEIIDRIVADWSEARKLAIETSVLEGNDGRLRGDELLVLAHTNDDVKRLN
EALRSVMTGEGALGEGRSFWAERGAREFAAGDRIIFLENARFLEPRAARLGPQYVKNGMLGTVVSTGDKR
GEVLLSVRLDNGRDVVIRENSYRNVDHGYAATIHKSQGATVERTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEARPEWGRKPRVDHAAGVTGELVETGQAKFRPNDEDADDSPYADVKTEDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVTIKVPVVDEQTGQKRFEEREVDRNVWTAKRIETAEARLERVERESHRP
DVFKQLVERLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDTLSRAAAGMEEEVSRRWAWIAEKREQVA
ELWERASMALGFAIERERRVSYNEERIETQSVSDADAGRARHLIAPTTEFARSAEEDARRAQLSSPAWKE
REAILRPLLEKIYRDPDAALSALNALSSDMSIEPRKLAEDLAAAPDRLGRLRGSDLIVDGRAARDERNAA
ITALTELLPLARAHATEFRQNGERFDLREQQRRAHMSLPIPALSKTAMARLVEIEAVRRQGGDDAYKTAF
ALATEDRSLVQEVKAVSEALAARFGWSAFTSKADAVAERNMVERMPEDLTSTRREKLARLFEAVKRFADE
QHLVERRDRSKIVAAASADQGMEAGKENAAVLPMLAAVTEFKTPVDEEARSRLLSAPLYRQQRAALAAVA
STIWRDPAGAVGKIEELLQKGFAGERIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKEAVQAMPEAAA
RLRALGSAYVNVLDAERQAITEERRRMTVAIPGLSKAAEEALMRLTAEAKNNGRKLSAPAASLDPGIRRE
FDAVSRALDERFGRNAIVRGEKDLINRIPPLQRRAFEAMQDRLKVLQQAVRMESSEQIISERRQRAVNRG
RGIGF
MAIMFVRAQVISRGVGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAPELVHEEVALPDWTPAWLRAAIDG
RTVAAASEALWNAVDVFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTQEGFGRKKVAVTGEDGAPLRVVTPDRPKGRIVYKLWAGDKETLQAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALSRRGAEMYFAPADLARRQEMADRLLADPTLLLKQ
LGNERSTFDERDIAKALHRYVDDPADFANIRARLMASDDLVMLKPQQVDAETGKASEPAVFTTREILRVE
YDMSQSARVLSERRGFAVSAESVAAAIENVETADPEKSFRLDAEQVDAVRHITGDSGIAAVVGLAGAGKS
TLLAAARLAWEHDGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELIWADGHETLHRGDVIVIDEAGMVS
SQQMARVLKLVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREDWAREASRLFARGE
IEKGLDAYAQQGHLIEAATRDEIIDRIVADWSEARKLAIETSVLEGNDGRLRGDELLVLAHTNDDVKRLN
EALRSVMTGEGALGEGRSFWAERGAREFAAGDRIIFLENARFLEPRAARLGPQYVKNGMLGTVVSTGDKR
GEVLLSVRLDNGRDVVIRENSYRNVDHGYAATIHKSQGATVERTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEARPEWGRKPRVDHAAGVTGELVETGQAKFRPNDEDADDSPYADVKTEDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVTIKVPVVDEQTGQKRFEEREVDRNVWTAKRIETAEARLERVERESHRP
DVFKQLVERLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDTLSRAAAGMEEEVSRRWAWIAEKREQVA
ELWERASMALGFAIERERRVSYNEERIETQSVSDADAGRARHLIAPTTEFARSAEEDARRAQLSSPAWKE
REAILRPLLEKIYRDPDAALSALNALSSDMSIEPRKLAEDLAAAPDRLGRLRGSDLIVDGRAARDERNAA
ITALTELLPLARAHATEFRQNGERFDLREQQRRAHMSLPIPALSKTAMARLVEIEAVRRQGGDDAYKTAF
ALATEDRSLVQEVKAVSEALAARFGWSAFTSKADAVAERNMVERMPEDLTSTRREKLARLFEAVKRFADE
QHLVERRDRSKIVAAASADQGMEAGKENAAVLPMLAAVTEFKTPVDEEARSRLLSAPLYRQQRAALAAVA
STIWRDPAGAVGKIEELLQKGFAGERIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKEAVQAMPEAAA
RLRALGSAYVNVLDAERQAITEERRRMTVAIPGLSKAAEEALMRLTAEAKNNGRKLSAPAASLDPGIRRE
FDAVSRALDERFGRNAIVRGEKDLINRIPPLQRRAFEAMQDRLKVLQQAVRMESSEQIISERRQRAVNRG
RGIGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 8032 | GenBank | NZ_CP022566 |
| Plasmid name | pSK02 | Incompatibility group | - |
| Plasmid size | 549819 bp | Coordinate of oriT [Strand] | 34724..34752 [-] |
| Host baterium | Rhizobium leguminosarum bv. viciae strain BIHB 1148 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |