Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107596 |
| Name | oriT_CCGM7|C |
| Organism | Sinorhizobium americanum CCGM7 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013054 (889159..889187 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_CCGM7|C
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file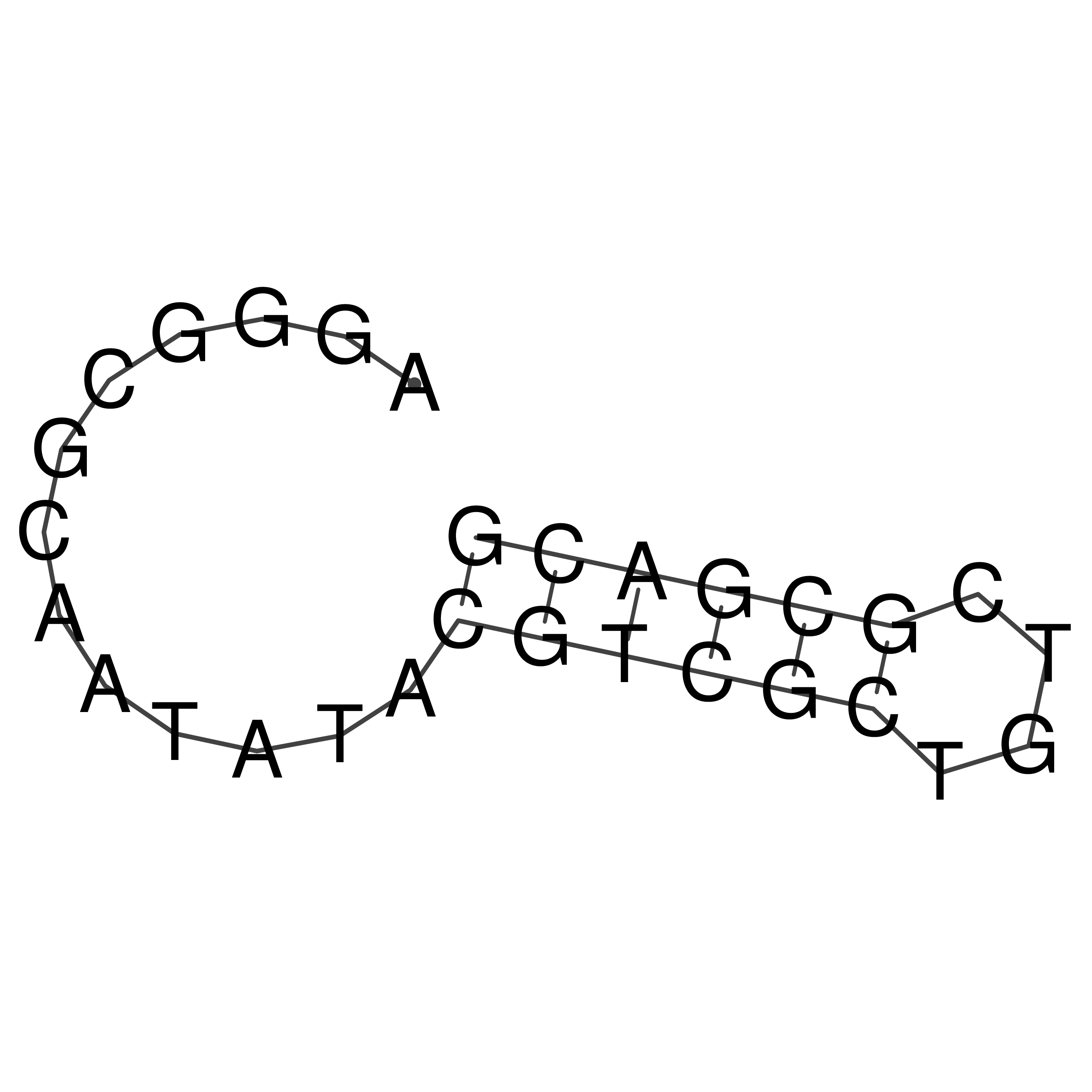
Relaxase
| ID | 5106 | GenBank | WP_037382860 |
| Name | traA_SAMCCGM7_RS25890_CCGM7|C |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170202.28 Da Isoelectric Point: 9.5559
>WP_037382860.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium americanum]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGSRELVHEELALPDQVPAWLKTATAG
QSVATASEALWNALDAFETRADAQLARELIIALPEELTRAENIGLVREFVRDSFTSKGMVADWVFHDKDG
NPHIHLMTTLRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIIYKLWAGDKEAMKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGVAQKHLGPERASLARKGAEMYFAPADLARRQEMADRLLNDPELLLKQ
LGNERSTFDEKDIAKALHRYVDDPVYFANIRARLMASDELVMLRPQQLDAETGKASEPAVFTTREILRIE
YDMARSAELLSQRKGFGVPGAEVAAAVQSIETADPAKPFKLEAEQVDAVSHVTGNSGIAAVVGLAGAGKS
TLLAAARVAWEREGRRVTGAALAGKAAEGLEDSSGIRSLTLASWELAWASGRETLHRGDVLVVDEAGMVS
SQQMGRVLKAVEDAGAKAVLVGDAMQLQPIQAGAAFRAISERVGFAELAGVRRQRDAWAREASRLLARGK
VEAGLDAYASRGRLVEAQTREEVIGRIISDWTDARKQAIEKSVSRGNEGRLRGDELLVLAHTNDDARKLN
GAVRKVMEDDGALSGGRAFQTARGAREFAVGDRIMFLENARFLEPRARRLGPQYVKNGMLGAVVSTGDKR
GDTLLSVRLDNGRHVVISEASYRNVDHGYAATIHKSQGSTFDRTFVLATGMMDQHLTYVAMTRHRERADL
YAAKEDFEAKPEWGREARIDHAAGVTGALVEVGMAKFRPDDEDAKESPYANIRTDDGTVHRVWGVSLPKA
IEDAGVCEGDTVTLRKDGVERVKVEVPVVDAETGQKRFEQREFDRNVWTAKQLETAETRQVRIERESHRP
ELFNQLVQRLSRSGAKTTTLDFESEAGYQAHAQDFARRRGIDTLAEVAAGVEEGVSRRLAWIAEKREQVA
KLWERASVALGFAIERERLVSYSEDRTELLSAGIPAGGRYLIPPTTEFARSVDEDARRALLSSPRWKERE
DILQPLLAKIYRDPDAALIHLNALASDAGIEPRTLASDLAKAPERLGRLRGSENLVAGRAARDERDIAMA
AFKELLPLVRAHATEFRRNADRFREREQTRRVYMSLSIPAISKQAMARLVEIEAVRSRGGDDAWKTAFAF
ATEDRSLVQEVKAVSDALTARFGWCAFTRKADAIAERNITERMPEDITDEWRGKLTRLFEAVRRFAEEQH
LAERKDRSKIVAGASVELGKEKMAVLPMLAAVTEFKTPVDDVARPRALAAQHYRHNRAALAETATRIWRD
PAGAVDRIEYLIVKGFAAERIAAAVVNDPAAYGALRGSDRLMDRMLAAGRERKDALQVVPEAAVHLRLLG
AAYVGAFDAERQAVTKERRLMAVAIPGLSPAAEEALRRLVSETKKKGSKLDVAAAALKPAIRQEFAAVSR
ALDERFGRNAILRGEKDVIKRVSPLQRRAFEAMHERLKVLQQTVRMQSSEQILSERRQRTINRARGVTW
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGSRELVHEELALPDQVPAWLKTATAG
QSVATASEALWNALDAFETRADAQLARELIIALPEELTRAENIGLVREFVRDSFTSKGMVADWVFHDKDG
NPHIHLMTTLRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIIYKLWAGDKEAMKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGVAQKHLGPERASLARKGAEMYFAPADLARRQEMADRLLNDPELLLKQ
LGNERSTFDEKDIAKALHRYVDDPVYFANIRARLMASDELVMLRPQQLDAETGKASEPAVFTTREILRIE
YDMARSAELLSQRKGFGVPGAEVAAAVQSIETADPAKPFKLEAEQVDAVSHVTGNSGIAAVVGLAGAGKS
TLLAAARVAWEREGRRVTGAALAGKAAEGLEDSSGIRSLTLASWELAWASGRETLHRGDVLVVDEAGMVS
SQQMGRVLKAVEDAGAKAVLVGDAMQLQPIQAGAAFRAISERVGFAELAGVRRQRDAWAREASRLLARGK
VEAGLDAYASRGRLVEAQTREEVIGRIISDWTDARKQAIEKSVSRGNEGRLRGDELLVLAHTNDDARKLN
GAVRKVMEDDGALSGGRAFQTARGAREFAVGDRIMFLENARFLEPRARRLGPQYVKNGMLGAVVSTGDKR
GDTLLSVRLDNGRHVVISEASYRNVDHGYAATIHKSQGSTFDRTFVLATGMMDQHLTYVAMTRHRERADL
YAAKEDFEAKPEWGREARIDHAAGVTGALVEVGMAKFRPDDEDAKESPYANIRTDDGTVHRVWGVSLPKA
IEDAGVCEGDTVTLRKDGVERVKVEVPVVDAETGQKRFEQREFDRNVWTAKQLETAETRQVRIERESHRP
ELFNQLVQRLSRSGAKTTTLDFESEAGYQAHAQDFARRRGIDTLAEVAAGVEEGVSRRLAWIAEKREQVA
KLWERASVALGFAIERERLVSYSEDRTELLSAGIPAGGRYLIPPTTEFARSVDEDARRALLSSPRWKERE
DILQPLLAKIYRDPDAALIHLNALASDAGIEPRTLASDLAKAPERLGRLRGSENLVAGRAARDERDIAMA
AFKELLPLVRAHATEFRRNADRFREREQTRRVYMSLSIPAISKQAMARLVEIEAVRSRGGDDAWKTAFAF
ATEDRSLVQEVKAVSDALTARFGWCAFTRKADAIAERNITERMPEDITDEWRGKLTRLFEAVRRFAEEQH
LAERKDRSKIVAGASVELGKEKMAVLPMLAAVTEFKTPVDDVARPRALAAQHYRHNRAALAETATRIWRD
PAGAVDRIEYLIVKGFAAERIAAAVVNDPAAYGALRGSDRLMDRMLAAGRERKDALQVVPEAAVHLRLLG
AAYVGAFDAERQAVTKERRLMAVAIPGLSPAAEEALRRLVSETKKKGSKLDVAAAALKPAIRQEFAAVSR
ALDERFGRNAILRGEKDVIKRVSPLQRRAFEAMHERLKVLQQTVRMQSSEQILSERRQRTINRARGVTW
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5329 | GenBank | WP_037382855 |
| Name | traG_SAMCCGM7_RS25875_CCGM7|C |
UniProt ID | _ |
| Length | 643 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 643 a.a. Molecular weight: 70739.48 Da Isoelectric Point: 9.4275
>WP_037382855.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium americanum]
MTPKAKPDPNLPLVLVPVAITAFAVYVVGWRWSEWAAGLSEKSAYWFLRASPVPTLSLGPLSGLLTVHML
PLHRRRPIAMASLVCFLTVAGYYALREYGRLAPAVESGAIRWDQALTYFDMVAVISAVLGFLAVAVAARI
STVVPDLVKRAKRGTLGDADWLPMSAAGKLFPPDGEIVVGERYRVDKELVHALPFDPNDTTTWGRGGKAP
LLTYGQNFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRTRVLGREVIVLDPT
NPIIGFNALDGIETSARQEEDIVGIAHMLLSDSARMESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQENSGSTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GSTFKSSDIVTARKDVFLNIPASILRSYPGIGRIVIGSLINAMVRADGGFKRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLVRHFGKDGATSWIEGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNGSRKSETVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNGAAK
ANRFVKARYEASD
MTPKAKPDPNLPLVLVPVAITAFAVYVVGWRWSEWAAGLSEKSAYWFLRASPVPTLSLGPLSGLLTVHML
PLHRRRPIAMASLVCFLTVAGYYALREYGRLAPAVESGAIRWDQALTYFDMVAVISAVLGFLAVAVAARI
STVVPDLVKRAKRGTLGDADWLPMSAAGKLFPPDGEIVVGERYRVDKELVHALPFDPNDTTTWGRGGKAP
LLTYGQNFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRTRVLGREVIVLDPT
NPIIGFNALDGIETSARQEEDIVGIAHMLLSDSARMESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQENSGSTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GSTFKSSDIVTARKDVFLNIPASILRSYPGIGRIVIGSLINAMVRADGGFKRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLVRHFGKDGATSWIEGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNGSRKSETVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNGAAK
ANRFVKARYEASD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5330 | GenBank | WP_037385045 |
| Name | tcpA_SAMCCGM7_RS30235_CCGM7|C |
UniProt ID | _ |
| Length | 928 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 928 a.a. Molecular weight: 100061.27 Da Isoelectric Point: 4.6314
>WP_037385045.1 DNA translocase FtsK [Sinorhizobium americanum]
MRIPRTNFSTAALHDADEADDFEHPYQGSPDGSHAQTTRASAQDHLFEAPDAPRRGAGHEADEMGYAGRA
ARLYGHGSAYAPAQVTASTRDPLPTLAEITGQTDGPSWESHFFLSPNVRFTRTPEREFMKRRAPAADESG
GEVEETGAEVAEPDVVAETAPADAPPATAEADTPAHTPSELLRVLMQQLPSWRPAQGRETALAESALDAE
PVVKEEVSVAEEPEGTPARAVRAVDVAEVILADIEPAPIVTGNESDAHLSYLSDHAFFEFMPLETVAVSQ
VAVEPVKELAKAPVQIATAPKPVALPPVEIRPQPPTAITSMFRVVECRPAATAVVAPGPAPVQIVNRSAE
EAVTAPPAPAPASVEPAESVKAEEDTPDLDVPVTKAAITMPAVVQRSTSTLPPVGRGERQQAGDTYEFPA
KELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEFEPAPGVKSSRVIGLAD
DIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESNDFQRTGCKLALCLGKTIGGEPVIAELA
KMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGIPHLLTPVVTDPKKAVMA
LKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGEPILATVQTGFEKGTGEPLFEQQEMDLTPMPYI
VVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIKANFPTRISFQVTSKIDS
RTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDLEVEHVVAHLKTQGRPEYLETVTADEEEPEEDQ
GAVFDKSAIAAEDGNELYEQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVERMEKDGLVGPANHVGKRE
IIHGNRDHAAKADDGEME
MRIPRTNFSTAALHDADEADDFEHPYQGSPDGSHAQTTRASAQDHLFEAPDAPRRGAGHEADEMGYAGRA
ARLYGHGSAYAPAQVTASTRDPLPTLAEITGQTDGPSWESHFFLSPNVRFTRTPEREFMKRRAPAADESG
GEVEETGAEVAEPDVVAETAPADAPPATAEADTPAHTPSELLRVLMQQLPSWRPAQGRETALAESALDAE
PVVKEEVSVAEEPEGTPARAVRAVDVAEVILADIEPAPIVTGNESDAHLSYLSDHAFFEFMPLETVAVSQ
VAVEPVKELAKAPVQIATAPKPVALPPVEIRPQPPTAITSMFRVVECRPAATAVVAPGPAPVQIVNRSAE
EAVTAPPAPAPASVEPAESVKAEEDTPDLDVPVTKAAITMPAVVQRSTSTLPPVGRGERQQAGDTYEFPA
KELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEFEPAPGVKSSRVIGLAD
DIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESNDFQRTGCKLALCLGKTIGGEPVIAELA
KMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGIPHLLTPVVTDPKKAVMA
LKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGEPILATVQTGFEKGTGEPLFEQQEMDLTPMPYI
VVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIKANFPTRISFQVTSKIDS
RTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDLEVEHVVAHLKTQGRPEYLETVTADEEEPEEDQ
GAVFDKSAIAAEDGNELYEQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVERMEKDGLVGPANHVGKRE
IIHGNRDHAAKADDGEME
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 858173..867728
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SAMCCGM7_RS25720 (SAMCCGM7_pC0851) | 853760..854881 | - | 1122 | WP_037382798 | Fic family protein | - |
| SAMCCGM7_RS34490 (SAMCCGM7_pC0852) | 855143..855268 | + | 126 | WP_281069543 | hypothetical protein | - |
| SAMCCGM7_RS25725 (SAMCCGM7_pC0853) | 855286..855477 | - | 192 | Protein_827 | type II toxin-antitoxin system VapC family toxin | - |
| SAMCCGM7_RS33295 (SAMCCGM7_pC0855) | 855925..856095 | + | 171 | WP_156878017 | hypothetical protein | - |
| SAMCCGM7_RS25730 (SAMCCGM7_pC0857) | 856569..856808 | - | 240 | WP_037382800 | hypothetical protein | - |
| SAMCCGM7_RS25735 (SAMCCGM7_pC0858) | 857163..857567 | - | 405 | WP_037382802 | MarR family transcriptional regulator | - |
| SAMCCGM7_RS34310 | 857664..858173 | + | 510 | WP_072597423 | hypothetical protein | - |
| SAMCCGM7_RS25745 (SAMCCGM7_pC0859) | 858173..858835 | + | 663 | WP_037382804 | lytic transglycosylase domain-containing protein | virB1 |
| SAMCCGM7_RS25750 (SAMCCGM7_pC0860) | 858832..859134 | + | 303 | WP_052035467 | TrbC/VirB2 family protein | virB2 |
| SAMCCGM7_RS25755 (SAMCCGM7_pC0861) | 859139..859477 | + | 339 | WP_037382805 | type IV secretion system protein VirB3 | virB3 |
| SAMCCGM7_RS25760 (SAMCCGM7_pC0862) | 859470..861836 | + | 2367 | WP_037382806 | VirB4 family type IV secretion system protein | virb4 |
| SAMCCGM7_RS25765 (SAMCCGM7_pC0863) | 861836..862531 | + | 696 | WP_037382808 | P-type DNA transfer protein VirB5 | virB5 |
| SAMCCGM7_RS25770 (SAMCCGM7_pC0864) | 862528..862761 | + | 234 | WP_037382810 | EexN family lipoprotein | - |
| SAMCCGM7_RS25775 (SAMCCGM7_pC0865) | 862767..863720 | + | 954 | WP_037382812 | type IV secretion system protein | virB6 |
| SAMCCGM7_RS25785 (SAMCCGM7_pC0866) | 864011..864682 | + | 672 | WP_037382814 | virB8 family protein | virB8 |
| SAMCCGM7_RS25790 (SAMCCGM7_pC0867) | 864694..865542 | + | 849 | WP_156878051 | P-type conjugative transfer protein VirB9 | virB9 |
| SAMCCGM7_RS25795 (SAMCCGM7_pC0868) | 865551..866726 | + | 1176 | WP_037382818 | type IV secretion system protein VirB10 | virB10 |
| SAMCCGM7_RS25800 (SAMCCGM7_pC0869) | 866739..867728 | + | 990 | WP_037382819 | P-type DNA transfer ATPase VirB11 | virB11 |
| SAMCCGM7_RS25805 | 867803..868090 | - | 288 | WP_037382820 | hypothetical protein | - |
| SAMCCGM7_RS25810 (SAMCCGM7_pC0871) | 868650..870467 | + | 1818 | WP_072597425 | ABC transporter ATP-binding protein | - |
| SAMCCGM7_RS25815 (SAMCCGM7_pC0872) | 870811..872067 | + | 1257 | WP_037382840 | hypothetical protein | - |
Host bacterium
| ID | 8031 | GenBank | NZ_CP013054 |
| Plasmid name | CCGM7|C | Incompatibility group | - |
| Plasmid size | 2249899 bp | Coordinate of oriT [Strand] | 889159..889187 [+] |
| Host baterium | Sinorhizobium americanum CCGM7 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | aioR/aoxR, aioS/aoxS, aioX/aoxX, arsC, actP |
| Degradation gene | benA, benB, benC |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |