Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107582 |
| Name | oriT_pRetKim5d |
| Organism | Rhizobium sp. Kim5 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021128 (241250..241278 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRetKim5d
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file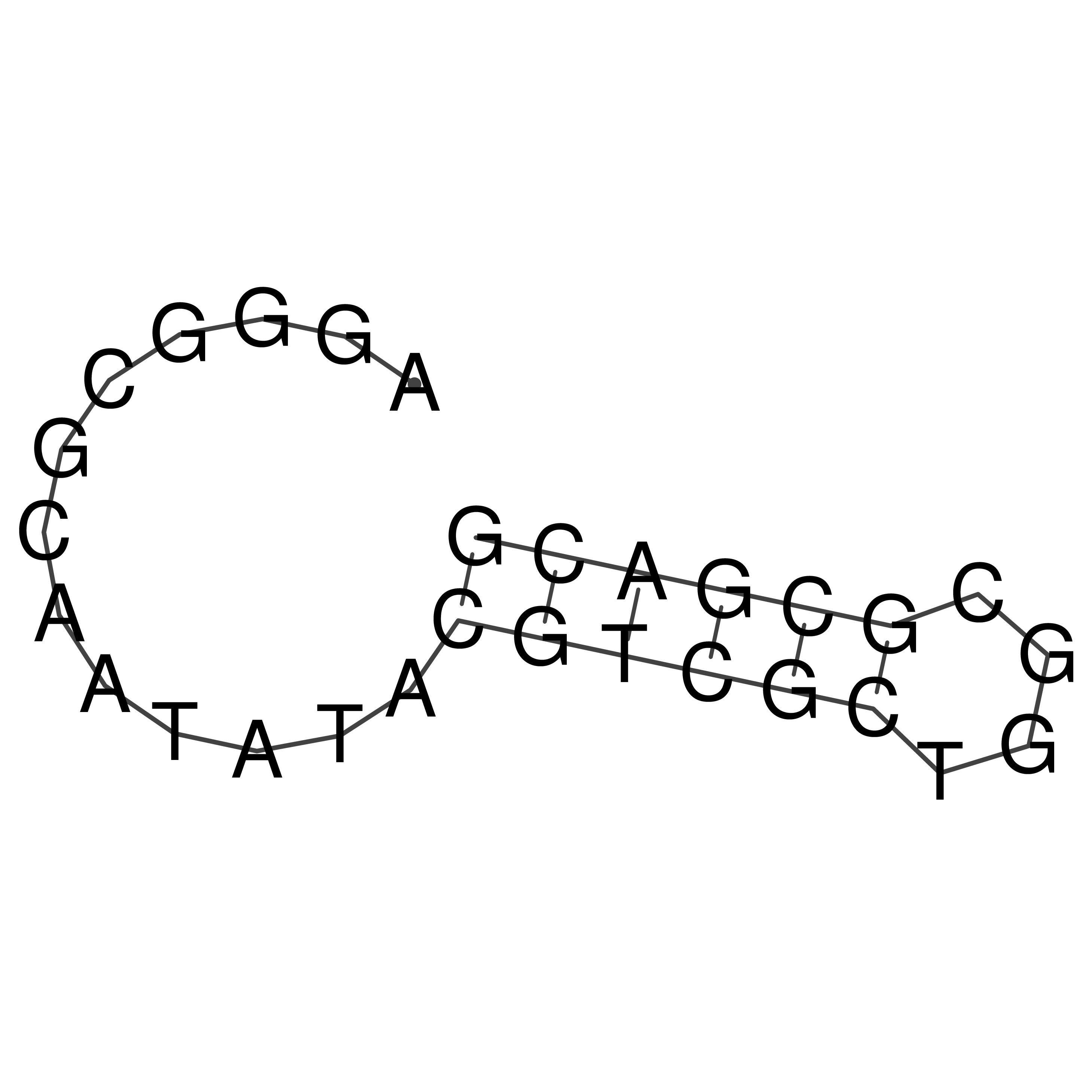
Relaxase
| ID | 5096 | GenBank | WP_086156714 |
| Name | traA_KIM5_RS31355_pRetKim5d |
UniProt ID | _ |
| Length | 1549 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1549 a.a. Molecular weight: 171076.43 Da Isoelectric Point: 6.6635
>WP_086156714.1 Ti-type conjugative transfer relaxase TraA [Rhizobium sp. Kim5]
MAIMFVRAQVISRGAGRNIVAAAAYRHRARMMDEQAGTSFSYRGGAGELVHEELALPEQTPAWLRAAIAG
HSVAGASEALWNAVDAFETRADAQLARELIIALPEELTRAENVALVRDFVRDNLTAKGMVADWVYHNKDG
NPHIHLMTTLRPLTEDGLGPKKVAVLGEDGKPLRVVTPDRPNGKIVYKVWGGDKEILKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGMAQKHLGPEKAALSRKGGERYFSPADLARRQEMADRLLADPELLLTR
LGNERSTFDERDIAKALHRYIDDPSDFANIRARLMASNELVMLKPQEIDAETGKAMEPAVFTIRDMLRIE
YDMAQSAKVLSERRGFAVSERHVNAAIESVETRDPQQRFRLEPEQVDAIRHVTGDDGIAAIVGLAGAGKS
TLLAAARLAWESENRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVVVIDEAGMVS
SQQMARVLDIVEQAEAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQHGRLVEAETRQEIVERIVADWTEARREAIGRSVAEGRAGNLRGDELLVLAHTNADVKRLN
EALRSVMTGEGALGESRSFRTERGSREFAAGDRIIFLENARFIEPRAGRLGPQYVKNGMLGTVVSTGDEK
GEVLLSVRLDNGSKVVFGGDSYSNVDHGYAATIHKSQGATVDRTFVLASGMMDRHLTYVSMTRHRDRADL
YAAKEDFAAKPEWGRKARVDHAAGVTGELIETGETKFRPDDEDAADSPYADIRTDDGAVHRLWGVSLPKA
LEDAFISEGDTVTLRKDGIERVKVQIPIVDEKTGQKRYEEMEVDRNVWTAKQIETADARQERIERESHRP
DVFEQLVERLSRSGAKTTTLDFESEAGYRAHAEDFARRRGLDHLSLAAAEMEESLSRRWAWIAAKREQVE
KLWERASVALGFAIERERRVAYNEERSQTIAEAASTDARYLIPPTISFARSIDEDARLAQLSSPAWKERE
ALLRPLLETIYRDPDAALVTLNALASDTSTEPRRLADNLAAAPDRLGRLRGSELIVEGRAARDERNVAMA
ALEDLLPLVRGHATEFRRNTERFELREQARRASMSLSVPALSDKAMARLMEIEAVRTQGGDDAYKTAFAL
AAKDRTVVQEIKAVSDAITARFGWSAFTSKADAIAERNMVERMPGDLTDERRGELSQLFEAVRRFAGEQH
LAERRDRSKVVAGASAVAGIEAAAGKDNVALLPMLAAVTEFKIPVDDEARSRILAAPLYRQQRAALAEAA
RMIWRDPANAVDKIEELLGKGFAADRIAAAVANDPAAYGALRGSDRLMDRMLAAGRERKEALQAVPEATA
RLRLLGAAYANILDAERQVITKERRRMAVAIPALSKAAEEALTRLTAAAQKDGRRLGVSAVALDPGIGRE
FADVSRALDERFGRNVLVRGDKDLVNRVPPAQRRAFEAMQERLKVLQQAVRLQSGEQILTERRQRTANRV
RGGDLSEPL
MAIMFVRAQVISRGAGRNIVAAAAYRHRARMMDEQAGTSFSYRGGAGELVHEELALPEQTPAWLRAAIAG
HSVAGASEALWNAVDAFETRADAQLARELIIALPEELTRAENVALVRDFVRDNLTAKGMVADWVYHNKDG
NPHIHLMTTLRPLTEDGLGPKKVAVLGEDGKPLRVVTPDRPNGKIVYKVWGGDKEILKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGMAQKHLGPEKAALSRKGGERYFSPADLARRQEMADRLLADPELLLTR
LGNERSTFDERDIAKALHRYIDDPSDFANIRARLMASNELVMLKPQEIDAETGKAMEPAVFTIRDMLRIE
YDMAQSAKVLSERRGFAVSERHVNAAIESVETRDPQQRFRLEPEQVDAIRHVTGDDGIAAIVGLAGAGKS
TLLAAARLAWESENRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVVVIDEAGMVS
SQQMARVLDIVEQAEAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQHGRLVEAETRQEIVERIVADWTEARREAIGRSVAEGRAGNLRGDELLVLAHTNADVKRLN
EALRSVMTGEGALGESRSFRTERGSREFAAGDRIIFLENARFIEPRAGRLGPQYVKNGMLGTVVSTGDEK
GEVLLSVRLDNGSKVVFGGDSYSNVDHGYAATIHKSQGATVDRTFVLASGMMDRHLTYVSMTRHRDRADL
YAAKEDFAAKPEWGRKARVDHAAGVTGELIETGETKFRPDDEDAADSPYADIRTDDGAVHRLWGVSLPKA
LEDAFISEGDTVTLRKDGIERVKVQIPIVDEKTGQKRYEEMEVDRNVWTAKQIETADARQERIERESHRP
DVFEQLVERLSRSGAKTTTLDFESEAGYRAHAEDFARRRGLDHLSLAAAEMEESLSRRWAWIAAKREQVE
KLWERASVALGFAIERERRVAYNEERSQTIAEAASTDARYLIPPTISFARSIDEDARLAQLSSPAWKERE
ALLRPLLETIYRDPDAALVTLNALASDTSTEPRRLADNLAAAPDRLGRLRGSELIVEGRAARDERNVAMA
ALEDLLPLVRGHATEFRRNTERFELREQARRASMSLSVPALSDKAMARLMEIEAVRTQGGDDAYKTAFAL
AAKDRTVVQEIKAVSDAITARFGWSAFTSKADAIAERNMVERMPGDLTDERRGELSQLFEAVRRFAGEQH
LAERRDRSKVVAGASAVAGIEAAAGKDNVALLPMLAAVTEFKIPVDDEARSRILAAPLYRQQRAALAEAA
RMIWRDPANAVDKIEELLGKGFAADRIAAAVANDPAAYGALRGSDRLMDRMLAAGRERKEALQAVPEATA
RLRLLGAAYANILDAERQVITKERRRMAVAIPALSKAAEEALTRLTAAAQKDGRRLGVSAVALDPGIGRE
FADVSRALDERFGRNVLVRGDKDLVNRVPPAQRRAFEAMQERLKVLQQAVRLQSGEQILTERRQRTANRV
RGGDLSEPL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5315 | GenBank | WP_086156713 |
| Name | traG_KIM5_RS31340_pRetKim5d |
UniProt ID | _ |
| Length | 638 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 638 a.a. Molecular weight: 70233.13 Da Isoelectric Point: 8.8072
>WP_086156713.1 Ti-type conjugative transfer system protein TraG [Rhizobium sp. Kim5]
MAVRRKLHPSLLLVLVPIVVTAITLAFISWRWQALAAGLSGRTEYWFLRAAPAPALLLGPAAGLLTVWAL
PLHQRRAVAMASLLCFMGVAGFYALREYSRLAPGVEHGIVTWDHALAYLDIVVVIGALTGFMISAVAARI
SVVVPEPVKRARLGTFGDADWLSMSAAAKLFPPDGEIVVGERYRVDKDIVHELPFDPVDRTTWGQGGKAA
LLTYGQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRMRALNREVMVLDPT
NPIVGFNVLAGIETSRQKEEDIVGVAHMLLSESLRFESSTGSYFQSQAHNLLTGLLAHVMLAPEYVGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSETAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFNPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMTQADGAFTRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGRDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKA
SSRNLGLDSRNGGTRKSESLNYHRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDAAAK
ANRFVKSV
MAVRRKLHPSLLLVLVPIVVTAITLAFISWRWQALAAGLSGRTEYWFLRAAPAPALLLGPAAGLLTVWAL
PLHQRRAVAMASLLCFMGVAGFYALREYSRLAPGVEHGIVTWDHALAYLDIVVVIGALTGFMISAVAARI
SVVVPEPVKRARLGTFGDADWLSMSAAAKLFPPDGEIVVGERYRVDKDIVHELPFDPVDRTTWGQGGKAA
LLTYGQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRMRALNREVMVLDPT
NPIVGFNVLAGIETSRQKEEDIVGVAHMLLSESLRFESSTGSYFQSQAHNLLTGLLAHVMLAPEYVGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSETAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFNPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMTQADGAFTRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGRDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKA
SSRNLGLDSRNGGTRKSESLNYHRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDAAAK
ANRFVKSV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 215223..234011
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KIM5_RS31165 (Kim5_PD00204) | 210537..211307 | + | 771 | WP_077994581 | 3-oxoacyl-ACP reductase family protein | - |
| KIM5_RS31170 (Kim5_PD00205) | 211486..212496 | + | 1011 | WP_077994582 | 2-keto-3-deoxygluconate permease | - |
| KIM5_RS35150 | 212534..212629 | + | 96 | Protein_211 | LysR family transcriptional regulator | - |
| KIM5_RS31180 | 212896..213312 | - | 417 | WP_348629256 | esterase | - |
| KIM5_RS31185 (Kim5_PD00206) | 213348..213791 | - | 444 | WP_077994590 | TA system VapC family ribonuclease toxin | - |
| KIM5_RS31190 (Kim5_PD00207) | 213788..214015 | - | 228 | WP_183758872 | CopG family transcriptional regulator | - |
| KIM5_RS31195 (Kim5_PD00208) | 214216..214581 | - | 366 | WP_077994592 | MarR family transcriptional regulator | - |
| KIM5_RS31200 (Kim5_PD00209) | 214680..215219 | + | 540 | WP_077994595 | hypothetical protein | - |
| KIM5_RS31205 (Kim5_PD00210) | 215223..215882 | + | 660 | WP_086156704 | transglycosylase SLT domain-containing protein | virB1 |
| KIM5_RS31210 (Kim5_PD00211) | 215879..216178 | + | 300 | WP_009986199 | TrbC/VirB2 family protein | virB2 |
| KIM5_RS31215 (Kim5_PD00212) | 216184..216522 | + | 339 | WP_009986201 | type IV secretion system protein VirB3 | virB3 |
| KIM5_RS31220 (Kim5_PD00213) | 216515..218881 | + | 2367 | WP_086156705 | VirB4 family type IV secretion system protein | virb4 |
| KIM5_RS31225 (Kim5_PD00214) | 218881..219579 | + | 699 | WP_009982580 | P-type DNA transfer protein VirB5 | virB5 |
| KIM5_RS31230 (Kim5_PD00215) | 219576..219809 | + | 234 | WP_010059082 | EexN family lipoprotein | - |
| KIM5_RS31235 (Kim5_PD00216) | 219813..220745 | + | 933 | WP_086156706 | type IV secretion system protein | virB6 |
| KIM5_RS31240 (Kim5_PD00217) | 220789..221073 | + | 285 | WP_086156707 | hypothetical protein | - |
| KIM5_RS31245 (Kim5_PD00218) | 221075..221746 | + | 672 | WP_009994555 | virB8 family protein | virB8 |
| KIM5_RS31250 (Kim5_PD00219) | 221743..222600 | + | 858 | WP_009994554 | P-type conjugative transfer protein VirB9 | virB9 |
| KIM5_RS31255 (Kim5_PD00220) | 222609..223787 | + | 1179 | WP_086156708 | type IV secretion system protein VirB10 | virB10 |
| KIM5_RS31260 (Kim5_PD00221) | 223789..224823 | + | 1035 | WP_245345117 | P-type DNA transfer ATPase VirB11 | virB11 |
| KIM5_RS31265 | 224797..225493 | - | 697 | Protein_229 | IS630 family transposase | - |
| KIM5_RS31270 (Kim5_PD00223) | 225643..226221 | - | 579 | WP_010056431 | PIN domain-containing protein | - |
| KIM5_RS31275 (Kim5_PD00224) | 226211..226657 | - | 447 | WP_040112772 | excisionase | - |
| KIM5_RS31280 (Kim5_PD00225) | 227116..229251 | + | 2136 | WP_086156709 | ParB/RepB/Spo0J family partition protein | - |
| KIM5_RS31285 (Kim5_PD00226) | 229363..230016 | + | 654 | WP_086156710 | hypothetical protein | - |
| KIM5_RS31290 (Kim5_PD00227) | 230147..231337 | + | 1191 | WP_040112775 | DUF932 domain-containing protein | - |
| KIM5_RS31295 (Kim5_PD00228) | 231404..232594 | - | 1191 | WP_040112776 | DUF1173 domain-containing protein | - |
| KIM5_RS31300 (Kim5_PD00229) | 232974..234011 | + | 1038 | WP_086156711 | toprim domain-containing protein | traP |
| KIM5_RS31305 (Kim5_PD00230) | 234321..235259 | + | 939 | WP_040112778 | DUF2493 domain-containing protein | - |
| KIM5_RS31310 (Kim5_PD00231) | 235410..235646 | + | 237 | WP_008530479 | type II toxin-antitoxin system VapB family antitoxin | - |
| KIM5_RS31315 (Kim5_PD00232) | 235646..236047 | + | 402 | WP_010059100 | type II toxin-antitoxin system VapC family toxin | - |
| KIM5_RS31320 (Kim5_PD00233) | 236184..236552 | - | 369 | WP_086156712 | WGR domain-containing protein | - |
| KIM5_RS31325 (Kim5_PD00234) | 236976..237299 | + | 324 | WP_008530485 | DUF736 family protein | - |
| KIM5_RS34465 | 237614..237781 | + | 168 | WP_164519071 | hypothetical protein | - |
| KIM5_RS31335 (Kim5_PD00237) | 238106..238588 | + | 483 | WP_009991774 | hypothetical protein | - |
Host bacterium
| ID | 8017 | GenBank | NZ_CP021128 |
| Plasmid name | pRetKim5d | Incompatibility group | - |
| Plasmid size | 684601 bp | Coordinate of oriT [Strand] | 241250..241278 [+] |
| Host baterium | Rhizobium sp. Kim5 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | hupE2 |
| Degradation gene | dxnH |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | - |