Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107579 |
| Name | oriT_pSymA |
| Organism | Sinorhizobium meliloti RU11/001 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021217 (1088650..1088706 [+], 57 nt) |
| oriT length | 57 nt |
| IRs (inverted repeats) | 4..9, 23..28 (GGAAAA..TTTTCC) |
| Location of nic site | 36..37 |
| Conserved sequence flanking the nic site |
TCCTGCCTCT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 57 nt
>oriT_pSymA
GCAGGAAAAGGGCGTAGCACATTTTTCCGTATCCTGCCTCTCCAAATTGTAAGGGGA
GCAGGAAAAGGGCGTAGCACATTTTTCCGTATCCTGCCTCTCCAAATTGTAAGGGGA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file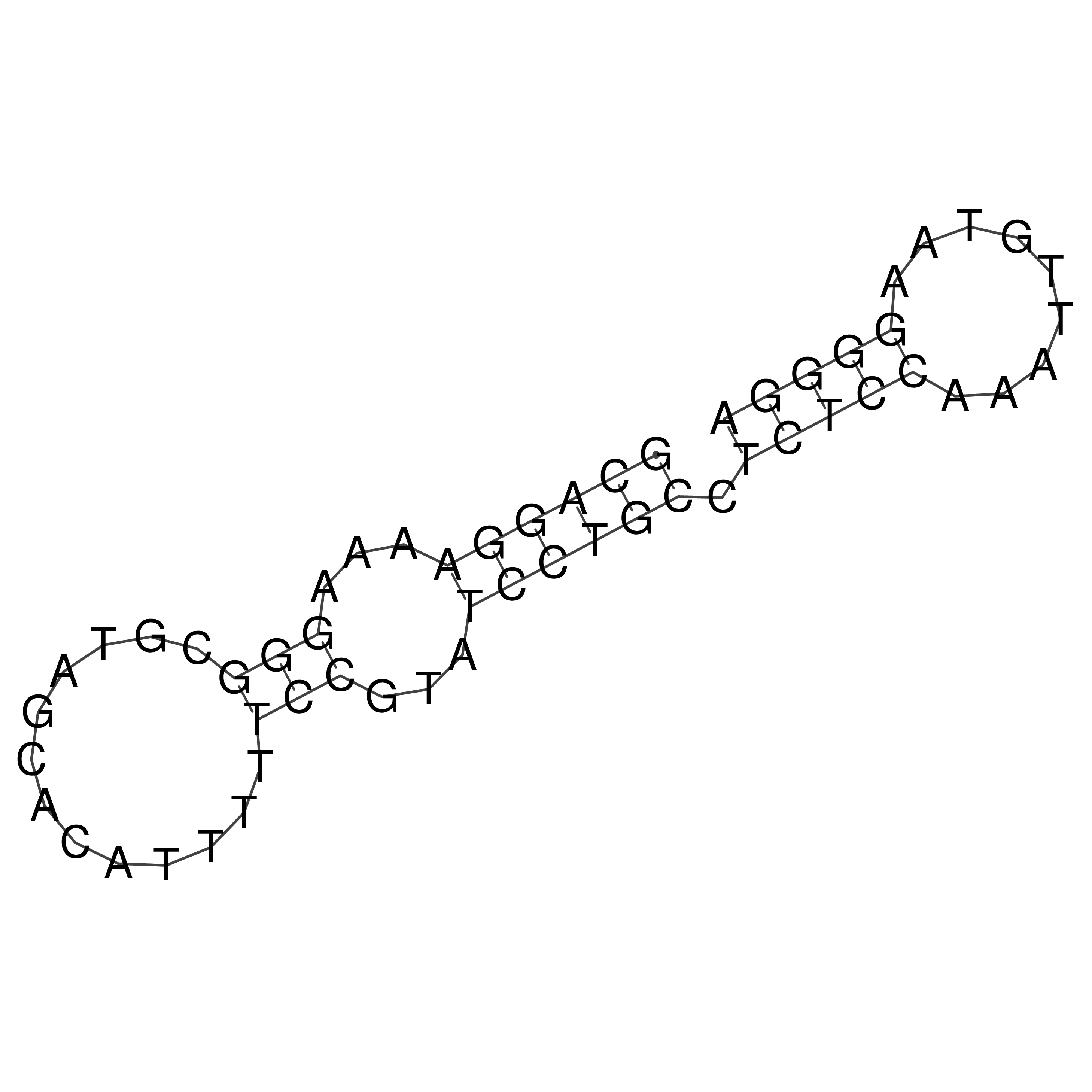
T4CP
| ID | 5311 | GenBank | WP_014531614 |
| Name | traG_SMRU11_RS05055_pSymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70270.30 Da Isoelectric Point: 9.4769
>WP_014531614.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKTHPSLLVILFPVAVTAAAVYVVGWRWPGLAAVMSGKTAYWFLRAAPVPALLFGPLAGLLAVWVL
PLHRRRPVAIASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAL
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVIEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAVP
MALKAKTHPSLLVILFPVAVTAAAVYVVGWRWPGLAAVMSGKTAYWFLRAAPVPALLFGPLAGLLAVWVL
PLHRRRPVAIASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAL
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVIEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5312 | GenBank | WP_014531067 |
| Name | t4cp2_SMRU11_RS08000_pSymA |
UniProt ID | _ |
| Length | 699 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 699 a.a. Molecular weight: 77789.72 Da Isoelectric Point: 9.6861
>WP_014531067.1 type IV secretory system conjugative DNA transfer family protein [Sinorhizobium meliloti]
MTKQLQAFYLLFCVGIAFVVWMLGYGLGLQLFYKDGRILETTITSNPFAPIQQFWHYKTSPALQKVALGS
LVPAMLVAGLVACIGLKPTSSPLGDAAFQDMASLRRGKWFRKQGHIFGRVGRNILRTRDDRHHLIIGPTR
SGKGAGYVIPNALMHEGSMIVTDLKGEVFKATAGYRRENGSQVFLFAPGSEKTSSYNPLDFIRPQRGNRT
TDIQNIASILVPENTASENSVWQATAQQVLAGAISYITESPFYKDRRNLAEVNSFFNSGVDLQALMKYIK
EKEPYLSKFTVESFNSYIALSERAAASALLDIQKAMRPFKNERIVAATNVTDMDLRAMKRRPISIYLAPN
ITDITLLRPLLTLFVQQVMDILTLEHDPNSLPVYFLLDEFRQLKRMDEIMTKLPYVAGYNIKLAFIIQDL
KNLDEIYGETSRHSLLGNCGYQLVLGANDQATAEYASRALGKRTIRYQSESRTIELMGLPRRTKVEQIRE
RDLMMPQEVRQMPENKMILLIEGQRPIFGEKLRFFQTQPFKSAEAFSQANIPRVPEVDYLSPKPVPATTP
EYAKGGEPSVEIPSSAIEKEEKPVAAAAIQSAAVKEEPAADDMPSTPAKRTVNRKALRPSAKATAANTGG
AEASPSLDAMEARIRAIEEGLKPKAAQLREVVEMKAEKLGDKSPTKRRNIMDIFSATVPDPVEVGVAAE
MTKQLQAFYLLFCVGIAFVVWMLGYGLGLQLFYKDGRILETTITSNPFAPIQQFWHYKTSPALQKVALGS
LVPAMLVAGLVACIGLKPTSSPLGDAAFQDMASLRRGKWFRKQGHIFGRVGRNILRTRDDRHHLIIGPTR
SGKGAGYVIPNALMHEGSMIVTDLKGEVFKATAGYRRENGSQVFLFAPGSEKTSSYNPLDFIRPQRGNRT
TDIQNIASILVPENTASENSVWQATAQQVLAGAISYITESPFYKDRRNLAEVNSFFNSGVDLQALMKYIK
EKEPYLSKFTVESFNSYIALSERAAASALLDIQKAMRPFKNERIVAATNVTDMDLRAMKRRPISIYLAPN
ITDITLLRPLLTLFVQQVMDILTLEHDPNSLPVYFLLDEFRQLKRMDEIMTKLPYVAGYNIKLAFIIQDL
KNLDEIYGETSRHSLLGNCGYQLVLGANDQATAEYASRALGKRTIRYQSESRTIELMGLPRRTKVEQIRE
RDLMMPQEVRQMPENKMILLIEGQRPIFGEKLRFFQTQPFKSAEAFSQANIPRVPEVDYLSPKPVPATTP
EYAKGGEPSVEIPSSAIEKEEKPVAAAAIQSAAVKEEPAADDMPSTPAKRTVNRKALRPSAKATAANTGG
AEASPSLDAMEARIRAIEEGLKPKAAQLREVVEMKAEKLGDKSPTKRRNIMDIFSATVPDPVEVGVAAE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 451249..460827
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SMRU11_RS04525 (SMRU11_04550) | 448142..448990 | + | 849 | WP_014531539 | alpha/beta fold hydrolase | - |
| SMRU11_RS04530 (SMRU11_04555) | 449040..449687 | + | 648 | WP_010967700 | dihydrofolate reductase family protein | - |
| SMRU11_RS04540 (SMRU11_04565) | 449808..450011 | + | 204 | WP_017266272 | hypothetical protein | - |
| SMRU11_RS04545 (SMRU11_04570) | 450272..450640 | - | 369 | WP_010967698 | transcriptional regulator | - |
| SMRU11_RS40950 (SMRU11_04575) | 450737..451245 | + | 509 | Protein_437 | hypothetical protein | - |
| SMRU11_RS04555 (SMRU11_04580) | 451249..451920 | + | 672 | WP_010967696 | lytic transglycosylase domain-containing protein | virB1 |
| SMRU11_RS04560 (SMRU11_04585) | 451917..452216 | + | 300 | WP_003526741 | TrbC/VirB2 family protein | virB2 |
| SMRU11_RS04565 (SMRU11_04590) | 452221..452562 | + | 342 | WP_003526740 | type IV secretion system protein VirB3 | virB3 |
| SMRU11_RS04570 (SMRU11_04595) | 452537..454933 | + | 2397 | WP_017266273 | VirB4 family type IV secretion system protein | virb4 |
| SMRU11_RS04575 (SMRU11_04600) | 454933..455634 | + | 702 | WP_014531544 | P-type DNA transfer protein VirB5 | virB5 |
| SMRU11_RS04580 (SMRU11_04605) | 455631..455846 | + | 216 | WP_029727813 | EexN family lipoprotein | - |
| SMRU11_RS04585 (SMRU11_04610) | 455869..456804 | + | 936 | WP_014531546 | type IV secretion system protein | virB6 |
| SMRU11_RS04590 (SMRU11_04615) | 456801..457115 | + | 315 | WP_013845458 | hypothetical protein | - |
| SMRU11_RS04595 (SMRU11_04620) | 457120..457791 | + | 672 | WP_014531547 | virB8 family protein | virB8 |
| SMRU11_RS04600 (SMRU11_04625) | 457791..458645 | + | 855 | WP_014531548 | P-type conjugative transfer protein VirB9 | virB9 |
| SMRU11_RS04605 (SMRU11_04630) | 458655..459827 | + | 1173 | WP_014531549 | type IV secretion system protein VirB10 | virB10 |
| SMRU11_RS04610 (SMRU11_04635) | 459838..460827 | + | 990 | WP_013845454 | P-type DNA transfer ATPase VirB11 | virB11 |
| SMRU11_RS04615 (SMRU11_04640) | 461138..462364 | - | 1227 | WP_013845453 | MFS transporter | - |
| SMRU11_RS04620 (SMRU11_04645) | 462401..463012 | - | 612 | WP_014531550 | TetR/AcrR family transcriptional regulator | - |
| SMRU11_RS04625 (SMRU11_04650) | 463539..463724 | + | 186 | WP_013845451 | periplasmic nitrate reductase, NapE protein | - |
| SMRU11_RS04630 (SMRU11_04655) | 463724..464224 | + | 501 | WP_013845450 | ferredoxin-type protein NapF | - |
| SMRU11_RS04635 (SMRU11_04660) | 464217..464504 | + | 288 | WP_013845449 | chaperone NapD | - |
Region 2: 1066116..1076039
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SMRU11_RS07905 (SMRU11_07930) | 1061285..1061620 | + | 336 | WP_013845322 | hypothetical protein | - |
| SMRU11_RS07910 (SMRU11_07935) | 1061702..1062118 | - | 417 | WP_014531084 | hypothetical protein | - |
| SMRU11_RS07915 (SMRU11_07940) | 1062142..1062735 | - | 594 | WP_026029870 | hypothetical protein | - |
| SMRU11_RS07925 (SMRU11_07950) | 1063140..1063841 | - | 702 | WP_014531082 | thermonuclease family protein | - |
| SMRU11_RS07930 (SMRU11_07955) | 1063838..1064374 | - | 537 | WP_014531081 | succinoglycan biosynthesis protein exoi | - |
| SMRU11_RS07935 (SMRU11_07960) | 1064371..1064979 | - | 609 | WP_014531080 | hypothetical protein | - |
| SMRU11_RS07940 (SMRU11_07965) | 1065202..1066104 | + | 903 | WP_029727939 | lytic transglycosylase domain-containing protein | - |
| SMRU11_RS07945 (SMRU11_07970) | 1066116..1066454 | + | 339 | WP_014531078 | TrbC/VirB2 family protein | virB2 |
| SMRU11_RS07950 (SMRU11_07975) | 1066458..1066757 | + | 300 | WP_014531077 | VirB3 family type IV secretion system protein | virB3 |
| SMRU11_RS07955 (SMRU11_07980) | 1066760..1069219 | + | 2460 | WP_014531076 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| SMRU11_RS07960 (SMRU11_07985) | 1069216..1070304 | + | 1089 | WP_014531075 | lytic transglycosylase domain-containing protein | - |
| SMRU11_RS07965 (SMRU11_07990) | 1070316..1071017 | + | 702 | WP_014531074 | type IV secretion system protein | - |
| SMRU11_RS07970 (SMRU11_07995) | 1071007..1071234 | + | 228 | WP_014531073 | hypothetical protein | - |
| SMRU11_RS07975 (SMRU11_08000) | 1071250..1072278 | + | 1029 | WP_014531072 | type IV secretion system protein | virB6 |
| SMRU11_RS07980 (SMRU11_08005) | 1072318..1073013 | + | 696 | WP_014531071 | type IV secretion system protein | virB8 |
| SMRU11_RS07985 (SMRU11_08010) | 1073010..1073819 | + | 810 | WP_014531070 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| SMRU11_RS07990 (SMRU11_08015) | 1073819..1075030 | + | 1212 | WP_014531069 | type IV secretion system protein VirB10 | virB10 |
| SMRU11_RS07995 (SMRU11_08020) | 1075011..1076039 | + | 1029 | WP_014531068 | P-type DNA transfer ATPase VirB11 | virB11 |
| SMRU11_RS08000 (SMRU11_08025) | 1076036..1078135 | + | 2100 | WP_014531067 | type IV secretory system conjugative DNA transfer family protein | - |
| SMRU11_RS08010 (SMRU11_08035) | 1078923..1080767 | + | 1845 | Protein_1076 | DUF3604 domain-containing protein | - |
Host bacterium
| ID | 8014 | GenBank | NZ_CP021217 |
| Plasmid name | pSymA | Incompatibility group | - |
| Plasmid size | 1613292 bp | Coordinate of oriT [Strand] | 1088650..1088706 [+] |
| Host baterium | Sinorhizobium meliloti RU11/001 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | ncrA |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB, fixN, fixO, fixG, fixS, nodM, nifN, nodD, nodA, nodB, nodC, nodI, nodJ, nodE, nodF, nodH, nifX, nifE, nifK, nifD, nifH, fixC, fixX, nifB, fixU |
| Anti-CRISPR | AcrIIA9 |