Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107577 |
| Name | oriT_pRlvD |
| Organism | Rhizobium leguminosarum bv. viciae strain UPM791 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP025510 (120599..120632 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file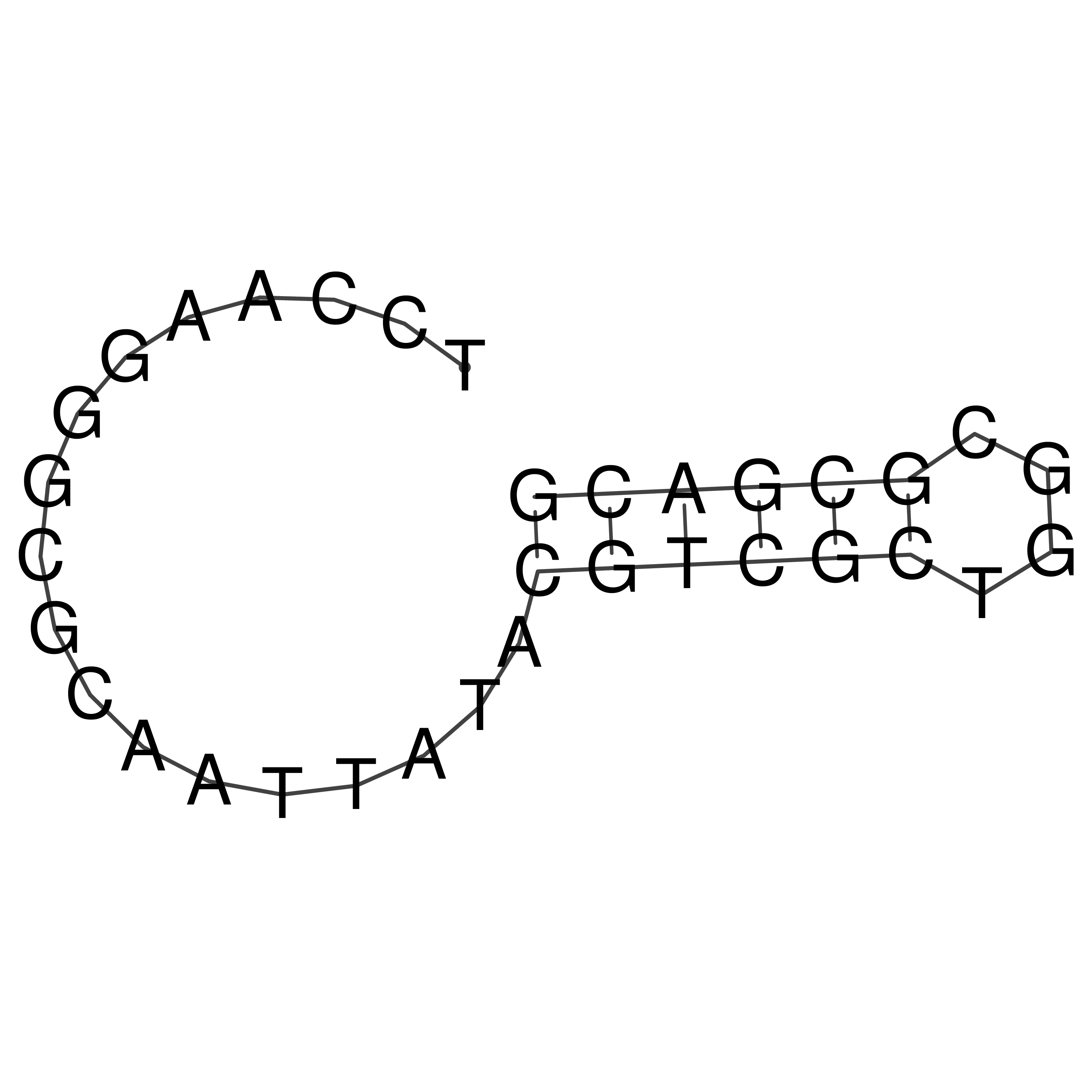
Relaxase
| ID | 5091 | GenBank | WP_104891287 |
| Name | traA_RLV_RS37660_pRlvD |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122922.11 Da Isoelectric Point: 9.6646
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEHNIALMRDFVERHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERVELQEQRRSENARRIQHRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFAMGIRTPEKYTTRDM
IRLEAQMANRAIWLSARATHGVREAVLQATFARHSRLSDEQRTAIEHITGATRIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLGDRCVFVLDEAGMVSSRQM
ALFVEAVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMLGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLAYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYE
KSALYRQALRFAEARGLHLMNVARTIAHDRLQWAVRQSSRLADLGARLAAIGKALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYANPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPEGFGALKGKTGLLASRADKQDLEKAVANVPALARNLERYLRERAEAELKHQTEERAVRLKV
SVDIPALSPSAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQLAAHERTTEALKQAETLRQTTSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5304 | GenBank | WP_245461641 |
| Name | t4cp2_RLV_RS37520_pRlvD |
UniProt ID | _ |
| Length | 541 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 541 a.a. Molecular weight: 59676.47 Da Isoelectric Point: 5.1482
MVSVKDYPAETYPGILDRLNLPYDTVVTQSFTPIDPISAQGKIARVRKQMRAAEDAAISLMAQLEEAEDD
VASGRVIFGDHHCSVAVFCDSLKELDGAMAYVARAMQDAGSAIVRENFSSRVVYFAQHPGNFAYRTRPAM
ISSQNFAQLCAMHGQSKGSPPDKCPWGDAVTIFPTARGETFRFNFHLPGKPGQRTVGHTLVLGQTGSGKT
LGTAFLISQAQRLGARTILFDKDNGFEMAVRAMGGRYSTVRMGDETGFNPIRAEADSRGAAWLTDWLAAL
AEYDGGKLNPLQIQALSDAVQANTSADPRLQNLEQFRSQLRATDDDGDLYNRLGRWDETGQFGWLFSGKG
LDPLAFDNRLTAFDLTEVFDNDVVRTAWLSYVFRRVERLVEDEHPTLLVLDEAWKLLDDPYFEARLKDWM
LTMRKKNVAVVLLTQRVSHITNSAAGDAILESAVTRLVYTSSFNTAAELAPLHLTPTESAFLQMSNVDNH
LVLLKSGSDSVVLDFDLVAIGKGIDVLGGGRGEKAIAGWRDRPDFQMEMLQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5305 | GenBank | WP_165505212 |
| Name | virD4_RLV_RS37870_pRlvD |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67296.23 Da Isoelectric Point: 7.8571
MDGSTTRKLNTLLFAFLLSLTVGLLFASIYASFYRHGISRETTNRFDIFSFWYETPLYVGYATPVFLRGL
TIAVSTAVLATMAGAIVILRKPHHHGTARWAARQELQRAGYIRPFRNITGPIFGKTVGPRWPGRYLTNGE
QPHSLVVAPTRAGKGVGVVIPTLLTFNGSILALDVKGELFELTSRARVTRGDRVFKFAPLDSAGRTHSYN
PVLDIVNMSPERRFSETRRLAVNLIVAKGKGSEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGE
KYKLFAQLAEETQNQEAQRIFDNMASNDTKILTSYTSVLGDGGLNLWADPLIKAATTTSDFSIYDLRRDP
TSIFLCVSPNDLEVIAPLIRLFFQQVVSILQRTLPKKDEIFEVLFLLDEFKHLGKLEAIETAVTTIAGYK
GRFMFIIQSLSALTGAYEESGKENFLSNTGIQIFMATADDETPNYISKAIGDYTFRARSTSFSQRNIFDT
NIQLSDQGAPLLRPEQVRLIDDDMEIVLIKGQPPLRLKKLKYYSDRVLKRIFDRQSGALPEPEPLHFATT
YVQAPATLLVAAESLPDEAKLEDAVQAALPDFDEVPVEGQGRCHLV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5306 | GenBank | WP_077988070 |
| Name | t4cp2_RLV_RS37960_pRlvD |
UniProt ID | _ |
| Length | 793 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 793 a.a. Molecular weight: 87501.45 Da Isoelectric Point: 6.2170
MAKGWCTVLERLATRAARSLALKRSEKGEMLVSGLPYLSAVDDVTLMLRDGDLMGSFVVEGINAETGDSF
HTVELCNAFSRFIAQQRSDVAYYVHRISVETRPTMPPVHGHPFNEEIDRRWQGYLQMVGLRDRVTVVTLV
IRPPRKTAKFLSLFGGRDVHSRHEVEARALRLDQMINTCMVGVGEAGPSRLTVSDGRWLGLLRTLIDGSG
GSLIPGTKFLPVADLLASSQITFQGPVFECEGTDPGRRRFGTIVSVKDYPAETYPGILDRLNLPYDTVVT
QSFTPIDPISAQGKIARVRKQMRAAEDAAISLMAQLEEAEDDVASGRVIFGDHHCSVAVFCDSLKELDGA
MAYVARAMQDAGSAIVRENFSSRVVYFAQHPGNFAYRTRPAMISSQNFAQLCAMHGQSKGSPPDKCPWGD
AVTIFPTARGETFRFNFHLPGKPGQRTVGHTLVLGQTGSGKTLGTAFLISQAQRLGARTILFDKDNGFEM
AVRAMGGRYSTVRMGDETGFNPIRAEADSRGAAWLTDWLAALAEYDGGKLNPLQIQALSDAVQANTSADP
RLQNLEQFRSQLRATDDDGDLYNRLGRWDETGQFGWLFSGKGLDPLAFDNRLTAFDLTEVFDNDVVRTAW
LSYVFRRVERLVEDEHPTLLVLDEAWKLLDDPYFEARLKDWMLTMRKKNVAVVLLTQRVSHITNSAAGDA
ILESAVTRLVYTSSFNTAAELAPLHLTPTESAFLQMSNVDNHLVLLKSGSDSVVLDFDLVAIGKGIDVLG
GGRGEKAIAGWRDRPDFQMEMLQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 82607..91785
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RLV_RS37495 (RLV_2107) | 77875..80563 | - | 2689 | Protein_91 | shikimate kinase | - |
| RLV_RS37500 (RLV_2108) | 80569..81319 | + | 751 | Protein_92 | IS6 family transposase | - |
| RLV_RS39420 (RLV_2109) | 81651..82484 | + | 834 | WP_245461638 | lytic transglycosylase domain-containing protein | - |
| RLV_RS37510 (RLV_2110) | 82607..82918 | + | 312 | WP_077988068 | TrbC/VirB2 family protein | virB2 |
| RLV_RS37515 | 83002..83280 | + | 279 | WP_104891276 | VirB3 family type IV secretion system protein | virB3 |
| RLV_RS39425 (RLV_2111) | 83339..84064 | + | 726 | WP_245461639 | hypothetical protein | virb4 |
| RLV_RS37520 (RLV_2112) | 84007..85632 | + | 1626 | WP_245461641 | DUF87 domain-containing protein | virb4 |
| RLV_RS37525 | 85763..86566 | + | 804 | WP_245315068 | type IV secretion system protein | - |
| RLV_RS39430 (RLV_2114) | 86607..87113 | + | 507 | WP_245461640 | hypothetical protein | virB6 |
| RLV_RS39435 | 87077..87715 | + | 639 | WP_245461642 | type IV secretion system protein | virB6 |
| RLV_RS37535 (RLV_2115) | 87928..88602 | + | 675 | WP_077988072 | type IV secretion system protein | virB8 |
| RLV_RS37540 (RLV_2116) | 88630..89412 | + | 783 | WP_077988073 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| RLV_RS37545 (RLV_2117) | 89612..90787 | + | 1176 | WP_104891313 | TrbI/VirB10 family protein | virB10 |
| RLV_RS37550 (RLV_2118) | 90784..91785 | + | 1002 | WP_104891277 | P-type DNA transfer ATPase VirB11 | virB11 |
| RLV_RS38420 | 92046..92246 | - | 201 | WP_131618399 | hypothetical protein | - |
| RLV_RS37555 (RLV_2119) | 92366..93088 | + | 723 | WP_104891278 | response regulator | - |
| RLV_RS37560 (RLV_2120) | 93247..93999 | + | 753 | WP_104891314 | IS6-like element ISRle39A family transposase | - |
Region 2: 181231..197179
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RLV_RS37905 (RLV_2194) | 179017..179769 | - | 753 | WP_104891314 | IS6-like element ISRle39A family transposase | - |
| RLV_RS37910 (RLV_2195) | 179928..180650 | - | 723 | WP_104891278 | response regulator | - |
| RLV_RS38435 | 180770..180970 | + | 201 | WP_131618399 | hypothetical protein | - |
| RLV_RS37915 (RLV_2197) | 181231..182232 | - | 1002 | WP_104891277 | P-type DNA transfer ATPase VirB11 | virB11 |
| RLV_RS37920 (RLV_2198) | 182229..183404 | - | 1176 | WP_104891313 | TrbI/VirB10 family protein | virB10 |
| RLV_RS37925 (RLV_2199) | 183604..184386 | - | 783 | WP_077988073 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| RLV_RS37930 (RLV_2200) | 184414..185088 | - | 675 | WP_077988072 | type IV secretion system protein | virB8 |
| RLV_RS37935 (RLV_2201) | 185181..186449 | - | 1269 | WP_104891304 | IS256 family transposase | - |
| RLV_RS37940 (RLV_2202) | 186552..186710 | + | 159 | Protein_190 | IS6 family transposase | - |
| RLV_RS37945 (RLV_2203) | 187042..187980 | + | 939 | WP_245461634 | lytic transglycosylase domain-containing protein | - |
| RLV_RS37950 (RLV_2204) | 187999..188310 | + | 312 | WP_077988068 | TrbC/VirB2 family protein | virB2 |
| RLV_RS37955 | 188394..188672 | + | 279 | WP_104891276 | VirB3 family type IV secretion system protein | virB3 |
| RLV_RS37960 (RLV_2205) | 188644..191025 | + | 2382 | WP_077988070 | DUF87 domain-containing protein | virb4 |
| RLV_RS37965 | 191156..191959 | + | 804 | WP_245315068 | type IV secretion system protein | - |
| RLV_RS37970 (RLV_2207) | 192000..193109 | + | 1110 | WP_245315069 | type IV secretion system protein | virB6 |
| RLV_RS37975 (RLV_2208) | 193322..193996 | + | 675 | WP_077988072 | type IV secretion system protein | virB8 |
| RLV_RS37980 (RLV_2209) | 194024..194806 | + | 783 | WP_077988073 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| RLV_RS37985 (RLV_2210) | 195006..196181 | + | 1176 | WP_104891313 | TrbI/VirB10 family protein | virB10 |
| RLV_RS37990 (RLV_2211) | 196178..197179 | + | 1002 | WP_104891277 | P-type DNA transfer ATPase VirB11 | virB11 |
| RLV_RS38440 | 197440..197640 | - | 201 | WP_131618399 | hypothetical protein | - |
| RLV_RS37995 (RLV_2212) | 197760..198482 | + | 723 | WP_104891278 | response regulator | - |
| RLV_RS39465 | 198565..198654 | + | 90 | Protein_203 | IS6 family transposase | - |
| RLV_RS38000 (RLV_2214) | 198716..199021 | - | 306 | Protein_204 | DDE-type integrase/transposase/recombinase | - |
| RLV_RS38005 (RLV_2215) | 199127..200293 | + | 1167 | WP_281060848 | IS256 family transposase | - |
| RLV_RS38010 (RLV_2216) | 200329..201012 | + | 684 | Protein_206 | IS5 family transposase | - |
Host bacterium
| ID | 8012 | GenBank | NZ_CP025510 |
| Plasmid name | pRlvD | Incompatibility group | - |
| Plasmid size | 217776 bp | Coordinate of oriT [Strand] | 120599..120632 [-] |
| Host baterium | Rhizobium leguminosarum bv. viciae strain UPM791 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |