Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107575 |
| Name | oriT_pRlvC |
| Organism | Rhizobium leguminosarum bv. viciae strain UPM791 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP025505 (384849..384889 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pRlvC
TCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file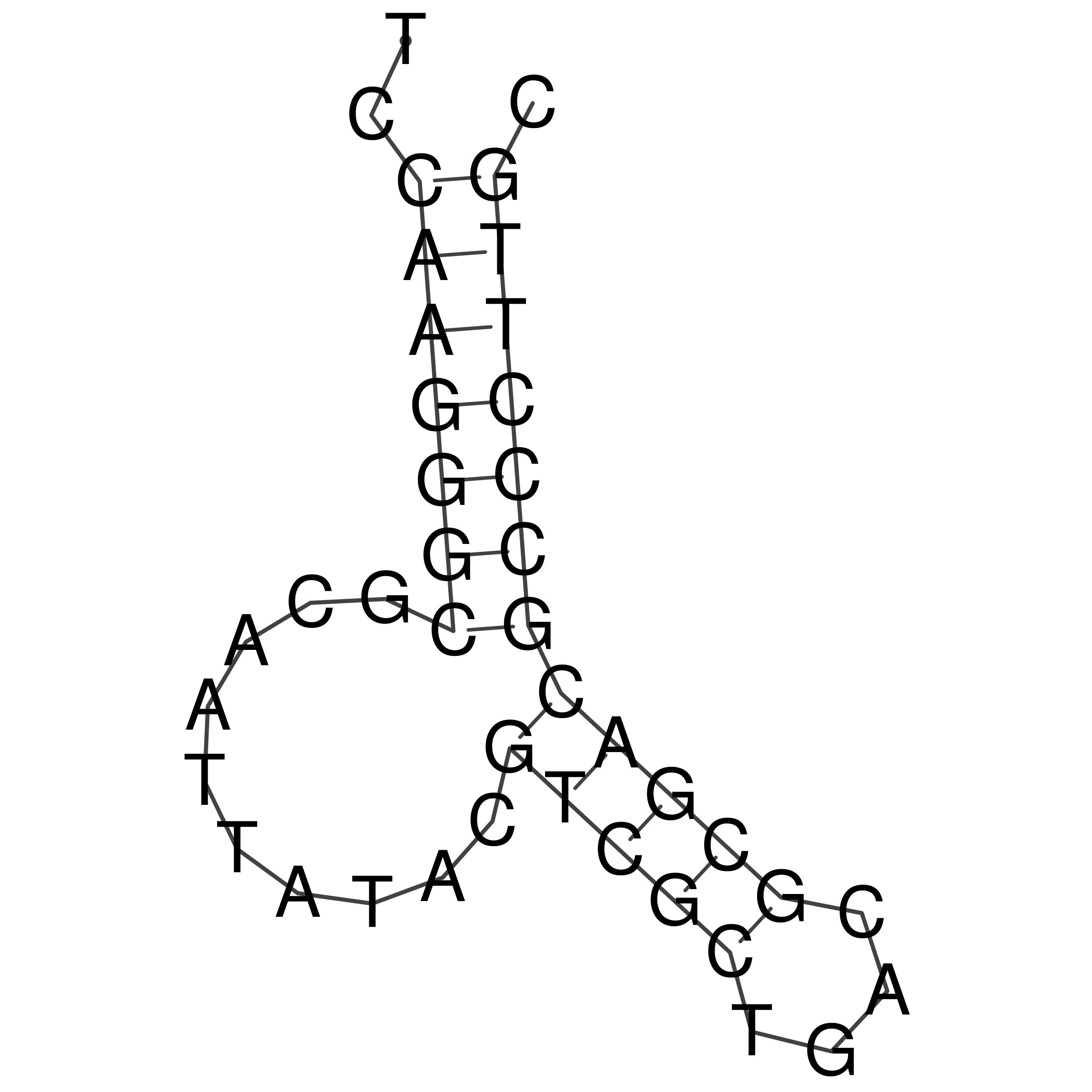
Relaxase
| ID | 5089 | GenBank | WP_018516732 |
| Name | traA_RLV_RS01765_pRlvC |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123258.54 Da Isoelectric Point: 9.9722
>WP_018516732.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTTALPLELTAEQNIALMRDFVVRHITAKGMVADWVYHDANGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDAKPIRNDAGKIVYELWAGSTEDFNAFRDGWFDCQNKHLALAG
LDIRIDGRSFEKQGIPLEPTIHLGVGTKAIARKAEQSDRKSETSTPKLERIELQEELRSENARRIQRRPE
IVLDLITKEKSVFDERDVAKVLYRYIDDVALFQSLMVRILRSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLQATFARHSRLSDEQRTAIEHVAGASRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRAGAKFVLVGDPEQLQPIEAGAAFRTIADRIGYAELETIYRQRDQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVENLIAAWDRDYDPSKTSLILSHLRRDVRKLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEIGEGEHRRLVMIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYFGSRSFAKSGGLIPILSRRNAKETSLDYE
KASFYGQALRFAEARGLHLMNVARTIARDRLEWTVRQKQKLADLGARLAAIGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDFSTRFQLVYAKPEAAFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGLRASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPSAKRTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAGKDTDG
KTFETITSGMTAGQKAEVQSAWNSMRTVQQLSSHERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTTALPLELTAEQNIALMRDFVVRHITAKGMVADWVYHDANGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDAKPIRNDAGKIVYELWAGSTEDFNAFRDGWFDCQNKHLALAG
LDIRIDGRSFEKQGIPLEPTIHLGVGTKAIARKAEQSDRKSETSTPKLERIELQEELRSENARRIQRRPE
IVLDLITKEKSVFDERDVAKVLYRYIDDVALFQSLMVRILRSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLQATFARHSRLSDEQRTAIEHVAGASRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRAGAKFVLVGDPEQLQPIEAGAAFRTIADRIGYAELETIYRQRDQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVENLIAAWDRDYDPSKTSLILSHLRRDVRKLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEIGEGEHRRLVMIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYFGSRSFAKSGGLIPILSRRNAKETSLDYE
KASFYGQALRFAEARGLHLMNVARTIARDRLEWTVRQKQKLADLGARLAAIGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDFSTRFQLVYAKPEAAFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGLRASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPSAKRTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAGKDTDG
KTFETITSGMTAGQKAEVQSAWNSMRTVQQLSSHERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5302 | GenBank | WP_018516735 |
| Name | traG_RLV_RS01750_pRlvC |
UniProt ID | _ |
| Length | 643 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 643 a.a. Molecular weight: 69508.71 Da Isoelectric Point: 9.2299
>WP_018516735.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVTGSSGTILIAAIRETMRLSAFMTVPADKTVWAFLDPATSIGASAALLCACFALRVALIGN
AAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGGK
SPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLDP
RKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGRTDEH
DQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAA
LVSGKKFATNEIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGYM
RIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTV
EIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGT
NRFHMVGDTPKPA
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVTGSSGTILIAAIRETMRLSAFMTVPADKTVWAFLDPATSIGASAALLCACFALRVALIGN
AAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGGK
SPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLDP
RKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGRTDEH
DQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAA
LVSGKKFATNEIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGYM
RIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTV
EIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGT
NRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5303 | GenBank | WP_018516716 |
| Name | t4cp2_RLV_RS01855_pRlvC |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91624.26 Da Isoelectric Point: 5.8767
>WP_018516716.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGTDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYADEESRKKSYADTVLFVFRNAVRELEQYFANTLSIRRMETRETLERGGERIARYDELLQFARFCTTG
ESHPIRLPDIPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYAANSPPLMQVASGSTSFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGSSLQPLTLAAGGDHYEIGGDNVEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQFLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLLK
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGTDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYADEESRKKSYADTVLFVFRNAVRELEQYFANTLSIRRMETRETLERGGERIARYDELLQFARFCTTG
ESHPIRLPDIPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYAANSPPLMQVASGSTSFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGSSLQPLTLAAGGDHYEIGGDNVEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQFLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 390021..404198
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RLV_RS01770 (RLV_2002) | 388285..388848 | + | 564 | WP_018516731 | conjugative transfer signal peptidase TraF | - |
| RLV_RS01775 (RLV_2003) | 388838..390004 | + | 1167 | WP_018516730 | conjugal transfer protein TraB | - |
| RLV_RS01780 (RLV_2004) | 390021..390638 | + | 618 | WP_018516729 | TraH family protein | virB1 |
| RLV_RS01785 (RLV_2005) | 390873..391742 | - | 870 | WP_018516728 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| RLV_RS01790 (RLV_2006) | 391735..392352 | - | 618 | WP_026242323 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| RLV_RS01800 (RLV_2007) | 392611..392883 | - | 273 | Protein_353 | DUF433 domain-containing protein | - |
| RLV_RS01805 (RLV_2008) | 392935..393147 | - | 213 | WP_018482035 | helix-turn-helix transcriptional regulator | - |
| RLV_RS01810 (RLV_2009) | 393353..393676 | + | 324 | WP_018482034 | transcriptional repressor TraM | - |
| RLV_RS01815 (RLV_2010) | 393680..394382 | - | 703 | Protein_356 | autoinducer binding domain-containing protein | - |
| RLV_RS01820 (RLV_2011) | 394694..395992 | - | 1299 | WP_018516723 | IncP-type conjugal transfer protein TrbI | virB10 |
| RLV_RS01825 (RLV_2012) | 396004..396444 | - | 441 | WP_018516722 | conjugal transfer protein TrbH | - |
| RLV_RS01830 (RLV_2013) | 396448..397257 | - | 810 | WP_026242322 | P-type conjugative transfer protein TrbG | virB9 |
| RLV_RS01835 (RLV_2014) | 397275..397937 | - | 663 | WP_018516720 | conjugal transfer protein TrbF | virB8 |
| RLV_RS01840 (RLV_2015) | 397958..399136 | - | 1179 | WP_018516719 | P-type conjugative transfer protein TrbL | virB6 |
| RLV_RS01845 | 399150..399326 | - | 177 | WP_018516718 | entry exclusion protein TrbK | - |
| RLV_RS01850 (RLV_2016) | 399320..400117 | - | 798 | WP_026242321 | P-type conjugative transfer protein TrbJ | virB5 |
| RLV_RS01855 (RLV_2017) | 400095..402545 | - | 2451 | WP_018516716 | conjugal transfer protein TrbE | virb4 |
| RLV_RS01860 (RLV_2018) | 402556..402855 | - | 300 | WP_018516715 | conjugal transfer protein TrbD | virB3 |
| RLV_RS01865 (RLV_2019) | 402848..403231 | - | 384 | WP_018516714 | TrbC/VirB2 family protein | virB2 |
| RLV_RS01870 (RLV_2020) | 403221..404198 | - | 978 | WP_018516713 | P-type conjugative transfer ATPase TrbB | virB11 |
| RLV_RS01875 (RLV_2021) | 404195..404833 | - | 639 | WP_018516712 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 8010 | GenBank | NZ_CP025505 |
| Plasmid name | pRlvC | Incompatibility group | - |
| Plasmid size | 405209 bp | Coordinate of oriT [Strand] | 384849..384889 [+] |
| Host baterium | Rhizobium leguminosarum bv. viciae strain UPM791 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | hupE |
| Degradation gene | - |
| Symbiosis gene | fixN, fixO, fixQ, nifN, nifE, nifD, nifH, fixA, fixB, fixC, fixX, nifB, nodJ, nodI, nodC, nodA, nodD, nodF, nodE, nodM |
| Anti-CRISPR | - |