Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107560 |
| Name | oriT_pSinA |
| Organism | Sinorhizobium sp. M14 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_021209 (88307..88343 [+], 37 nt) |
| oriT length | 37 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 37 nt
>oriT_pSinA
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACG
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file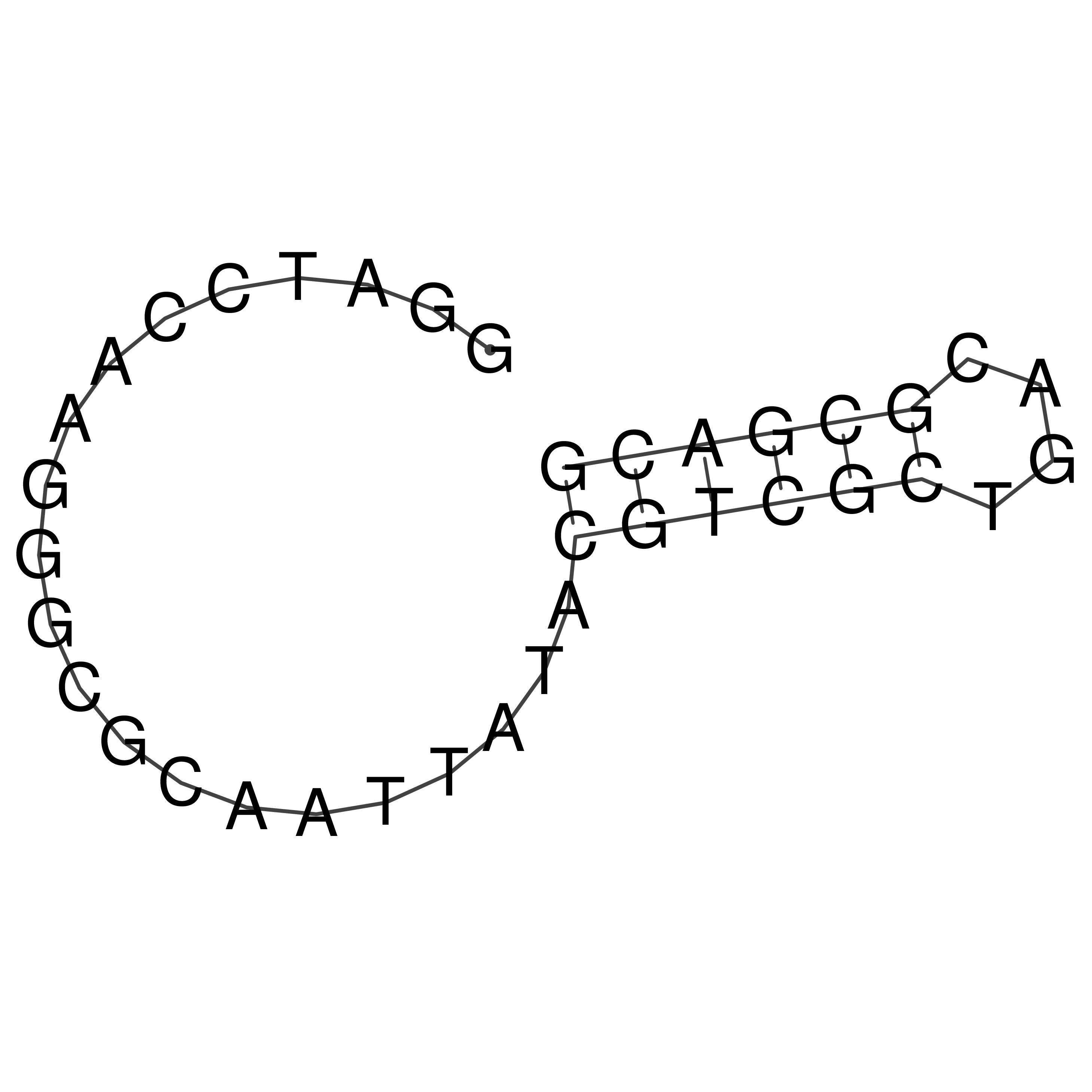
Relaxase
| ID | 5073 | GenBank | WP_015647745 |
| Name | traA_HS899_RS00435_pSinA |
UniProt ID | R4IL57 |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 123035.13 Da Isoelectric Point: 9.7753
>WP_015647745.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Sinorhizobium/Ensifer group]
MAVPHFSVSIVARGSGRSAVLSAAYRHCARMDYEREARTIDYTGKQGLLHEEFVIPADAPDWLRTMIADR
SVSGASEAFWNKVEAFEKRVDAQLAKDITIALPIELSSEQNIALMQDFVAEHLTTKGMVADWVYHDAPGN
PHVHLMTTLRPLSEDGFGAKKVTVRGSDGQPMRNDAGKIIYELWAGGAEDFNAFRDGWFAVQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHIGVGATAIERKSEMENRLATAGSRKLERIELQEERRAENVRRIQRNPG
IVLDLITRERSVFDNQDVAKILHRYVDDAALFQSLMARIMQHLDVLRLDCERINFTSGVRTPARYTTREM
IRLEAEMANRSIWLSQRSSHGVRQKIIEAVFERHKRLSDEQKTAIEHVAGQERIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGAALAGKAAEGLEKEAGILSRTLSSWELRWAQGRDQLDDKTVMVLDEAGMVSSKQM
ALLVEEATVRGAKLILIGDPEQLQPIEAGAAFRAITDRIGYAELETIYRQREQWMRDASLDLARGNIAKA
VESYSANGRMMGSTLKSQAVENLISDWNREYDPAHSSLILAHLRRDVRMLNTLARAKLVERGLVDDGHAF
KTEDGVRHFAAGDRIVFLKNESSLGVKNGMLAKVVEATPGRIVAEIGEGEHRKAISVEQRFYNNVDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDIGVYYGARSFAKAGGLAELLSRTNSKETTLDYE
KGAFYRAALRFADARGLHLVNVARTLVRDRLDWTVRQKQKLFDLTARLATIRAQLGLKGPNTQVTSKPDM
EAKPMVSGITTFPKSIDHAVEDRLVADPGLKKQWQEVTTRFIQVFAEPETAFKAVNVDAMLKDPARAQTT
LAKIAAEPERFGALKGKTGIFAGANEKAARDTALVNAPALARNLERYVTARVEAERKHEAQERAIRLKVS
IDIPALSPSARQTLERIRDAIDRNDLSAGLEYALADRNVKAELEGFAKAVSERFGERSLLPISAKDANGE
TFHKITAGMTPSQKSEVKSAWNSMRTVQQLAAHERTTLALKQAETARQTQTKGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCARMDYEREARTIDYTGKQGLLHEEFVIPADAPDWLRTMIADR
SVSGASEAFWNKVEAFEKRVDAQLAKDITIALPIELSSEQNIALMQDFVAEHLTTKGMVADWVYHDAPGN
PHVHLMTTLRPLSEDGFGAKKVTVRGSDGQPMRNDAGKIIYELWAGGAEDFNAFRDGWFAVQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHIGVGATAIERKSEMENRLATAGSRKLERIELQEERRAENVRRIQRNPG
IVLDLITRERSVFDNQDVAKILHRYVDDAALFQSLMARIMQHLDVLRLDCERINFTSGVRTPARYTTREM
IRLEAEMANRSIWLSQRSSHGVRQKIIEAVFERHKRLSDEQKTAIEHVAGQERIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGAALAGKAAEGLEKEAGILSRTLSSWELRWAQGRDQLDDKTVMVLDEAGMVSSKQM
ALLVEEATVRGAKLILIGDPEQLQPIEAGAAFRAITDRIGYAELETIYRQREQWMRDASLDLARGNIAKA
VESYSANGRMMGSTLKSQAVENLISDWNREYDPAHSSLILAHLRRDVRMLNTLARAKLVERGLVDDGHAF
KTEDGVRHFAAGDRIVFLKNESSLGVKNGMLAKVVEATPGRIVAEIGEGEHRKAISVEQRFYNNVDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDIGVYYGARSFAKAGGLAELLSRTNSKETTLDYE
KGAFYRAALRFADARGLHLVNVARTLVRDRLDWTVRQKQKLFDLTARLATIRAQLGLKGPNTQVTSKPDM
EAKPMVSGITTFPKSIDHAVEDRLVADPGLKKQWQEVTTRFIQVFAEPETAFKAVNVDAMLKDPARAQTT
LAKIAAEPERFGALKGKTGIFAGANEKAARDTALVNAPALARNLERYVTARVEAERKHEAQERAIRLKVS
IDIPALSPSARQTLERIRDAIDRNDLSAGLEYALADRNVKAELEGFAKAVSERFGERSLLPISAKDANGE
TFHKITAGMTPSQKSEVKSAWNSMRTVQQLAAHERTTLALKQAETARQTQTKGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | R4IL57 |
T4CP
| ID | 5289 | GenBank | WP_015647742 |
| Name | traG_HS899_RS00420_pSinA |
UniProt ID | _ |
| Length | 645 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 645 a.a. Molecular weight: 69657.82 Da Isoelectric Point: 9.3417
>WP_015647742.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Sinorhizobium/Ensifer group]
MTANRILLAVVPPLVMIAIALMLPGIEQWLASFGKTAEAKLTLGRIGLSLPYIASAAIGLIFLLSAQGSV
NIKTAGWGVAAGSGAVIAIAVVREGMRLSQFAGQAPAARSILSYVDPATMTGAAAALFSGMFALRVAVIG
NAAFSRSEPKRIRGKRALHGEAEWMKLPEAEKLFADGGGIVIGERYRVDRDSTAALSFRADQPETWGTGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVKAHRIKAGRDVIVLD
PKDPDIGFNALDWIGQHGGTKEEDIAAVASWIMSESGRVSGVRDDFFRASGLQLLTAIIADVCLSGHTEK
KNQTLRTVRMNLSEPEPKLRARLQEIYDNSESDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYPNYA
GLVSGSTFSTDDIASGTTDVFVALDLKTLETHGGLARVIIGSFLNAIYNRDGAMPGRALFLLDEVARLGF
MRILETARDAGRKYGITLLMIYQSIGQMRETYGGRDATSKWFESASWISFSAINDPETADYISRRCGTTT
VEIDQVSRSFQARGSSRTRSKQLASRPLIQPHEVLRMRADEQIVFTGGNAPLRCGRAMWFRRADMKSCVG
DNRFHGNRGPLEIKV
MTANRILLAVVPPLVMIAIALMLPGIEQWLASFGKTAEAKLTLGRIGLSLPYIASAAIGLIFLLSAQGSV
NIKTAGWGVAAGSGAVIAIAVVREGMRLSQFAGQAPAARSILSYVDPATMTGAAAALFSGMFALRVAVIG
NAAFSRSEPKRIRGKRALHGEAEWMKLPEAEKLFADGGGIVIGERYRVDRDSTAALSFRADQPETWGTGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVKAHRIKAGRDVIVLD
PKDPDIGFNALDWIGQHGGTKEEDIAAVASWIMSESGRVSGVRDDFFRASGLQLLTAIIADVCLSGHTEK
KNQTLRTVRMNLSEPEPKLRARLQEIYDNSESDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYPNYA
GLVSGSTFSTDDIASGTTDVFVALDLKTLETHGGLARVIIGSFLNAIYNRDGAMPGRALFLLDEVARLGF
MRILETARDAGRKYGITLLMIYQSIGQMRETYGGRDATSKWFESASWISFSAINDPETADYISRRCGTTT
VEIDQVSRSFQARGSSRTRSKQLASRPLIQPHEVLRMRADEQIVFTGGNAPLRCGRAMWFRRADMKSCVG
DNRFHGNRGPLEIKV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5290 | GenBank | WP_015647761 |
| Name | t4cp2_HS899_RS00525_pSinA |
UniProt ID | _ |
| Length | 811 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 811 a.a. Molecular weight: 91066.53 Da Isoelectric Point: 5.7391
>WP_015647761.1 MULTISPECIES: conjugal transfer protein TrbE [Sinorhizobium/Ensifer group]
MVALCSLRPSAPSFADLVPYAGLVDNGTILLKDGSLMAGWYFAGPDSESATDFERNDLSRQINTILSRLG
SGWMIQVEAVRIPTTDYPSAERSHFPDAVTRMIDDERRKHFEQERGHFESRHALVLTYRPPERRRSGVTR
YIYSDTESRTATYADTVLDSFRRSTREIEQYLGNVISIERMRTREVPERGGTRIARYDELFQFIRFCITG
ENHPVCLPEIPMYLDWLATAELEHGLTPKVENRFLSAIAIDGLPAESWPGILNSLDLLPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIIL
FDESREKLADKAESVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLIPLNSVWS
GSPLAPCPFYPPDAPPLTQVASGSSPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYKGAQVFA
FDKGNSILPLTLAVDGNHYEIGGEAGEGARLAFCPLSDLSTETDRAWASEWIETLVSLQGVTVNPDHRNA
ISRQIGLMAGAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDSLTLGQFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAERS
GIIDVLKESCPTKICLPNGAARESGTREFYERIGFNPRQIEIVAAAIPKREYYVTSPDGRRLFDMALGPL
TLSFVGASGKADLARIRELLSTYGKDWPTQWLNERGINRYA
MVALCSLRPSAPSFADLVPYAGLVDNGTILLKDGSLMAGWYFAGPDSESATDFERNDLSRQINTILSRLG
SGWMIQVEAVRIPTTDYPSAERSHFPDAVTRMIDDERRKHFEQERGHFESRHALVLTYRPPERRRSGVTR
YIYSDTESRTATYADTVLDSFRRSTREIEQYLGNVISIERMRTREVPERGGTRIARYDELFQFIRFCITG
ENHPVCLPEIPMYLDWLATAELEHGLTPKVENRFLSAIAIDGLPAESWPGILNSLDLLPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIIL
FDESREKLADKAESVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLIPLNSVWS
GSPLAPCPFYPPDAPPLTQVASGSSPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYKGAQVFA
FDKGNSILPLTLAVDGNHYEIGGEAGEGARLAFCPLSDLSTETDRAWASEWIETLVSLQGVTVNPDHRNA
ISRQIGLMAGAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDSLTLGQFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAERS
GIIDVLKESCPTKICLPNGAARESGTREFYERIGFNPRQIEIVAAAIPKREYYVTSPDGRRLFDMALGPL
TLSFVGASGKADLARIRELLSTYGKDWPTQWLNERGINRYA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 93482..108825
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HS899_RS00440 (ST430451_p045) | 91742..92308 | + | 567 | WP_015647746 | conjugative transfer signal peptidase TraF | - |
| HS899_RS00445 (ST430451_p044) | 92298..93464 | + | 1167 | WP_015647747 | conjugal transfer protein TraB | - |
| HS899_RS00450 (ST430451_p043) | 93482..94093 | + | 612 | WP_015647748 | TraH family protein | virB1 |
| HS899_RS00455 (ST430451_p042) | 94126..94746 | - | 621 | WP_015647749 | RES family NAD+ phosphorylase | - |
| HS899_RS00460 (ST430451_p088) | 94747..95502 | - | 756 | WP_015647750 | hypothetical protein | - |
| HS899_RS00465 | 95637..95888 | - | 252 | WP_115422184 | type II toxin-antitoxin system VapB family antitoxin | - |
| HS899_RS00470 (ST430451_p089) | 96193..96897 | + | 705 | WP_015647751 | transcriptional regulator TraR | - |
| HS899_RS00475 (ST430451_p090) | 96912..97211 | - | 300 | WP_015647752 | transcriptional repressor TraM | - |
| HS899_RS00480 (ST430451_p091) | 97677..98483 | + | 807 | WP_343215831 | LuxR family transcriptional regulator | - |
| HS899_RS00485 (ST430451_p092) | 98601..99263 | + | 663 | WP_015647754 | acyl-homoserine-lactone synthase | - |
| HS899_RS00490 (ST430451_p093) | 99303..100598 | - | 1296 | WP_015647755 | IncP-type conjugal transfer protein TrbI | virB10 |
| HS899_RS00495 (ST430451_p094) | 100611..101054 | - | 444 | WP_015647756 | conjugal transfer protein TrbH | - |
| HS899_RS00500 (ST430451_p095) | 101058..101888 | - | 831 | WP_015647757 | P-type conjugative transfer protein TrbG | virB9 |
| HS899_RS00505 (ST430451_p096) | 101904..102566 | - | 663 | WP_015647758 | conjugal transfer protein TrbF | virB8 |
| HS899_RS00510 (ST430451_p097) | 102588..103769 | - | 1182 | WP_015647759 | P-type conjugative transfer protein TrbL | virB6 |
| HS899_RS00515 | 103774..103950 | - | 177 | WP_115422182 | entry exclusion protein TrbK | - |
| HS899_RS00520 (ST430451_p098) | 103944..104747 | - | 804 | WP_015647760 | P-type conjugative transfer protein TrbJ | virB5 |
| HS899_RS00525 (ST430451_p099) | 104740..107175 | - | 2436 | WP_015647761 | conjugal transfer protein TrbE | virb4 |
| HS899_RS00530 (ST430451_p100) | 107186..107485 | - | 300 | WP_015647762 | conjugal transfer protein TrbD | virB3 |
| HS899_RS00535 (ST430451_p101) | 107478..107870 | - | 393 | WP_015647763 | TrbC/VirB2 family protein | virB2 |
| HS899_RS00540 (ST430451_p102) | 107860..108825 | - | 966 | WP_015647764 | P-type conjugative transfer ATPase TrbB | virB11 |
Host bacterium
| ID | 7995 | GenBank | NC_021209 |
| Plasmid name | pSinA | Incompatibility group | - |
| Plasmid size | 108938 bp | Coordinate of oriT [Strand] | 88307..88343 [+] |
| Host baterium | Sinorhizobium sp. M14 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | aioR/aoxR, aioS/aoxS, aioX/aoxX, arsC, arsH |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |